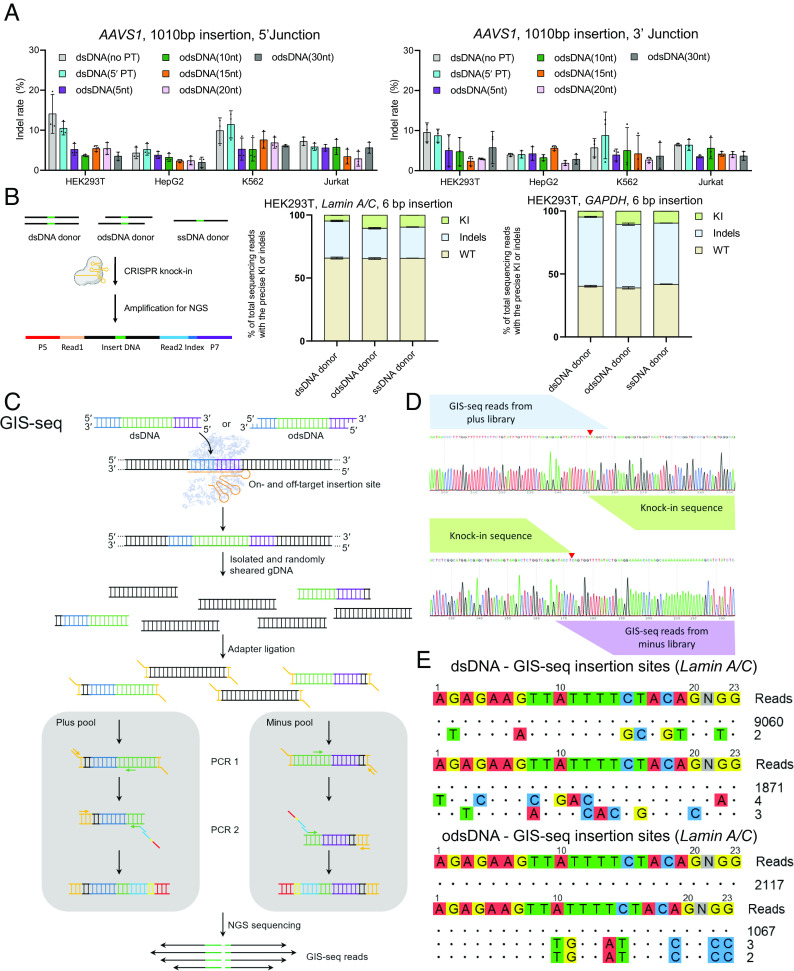
Fig. 3.
KI with odsDNA donors exhibited enhanced HDR frequencies with low off-target integration. (A) Comparative analyses for the on-target indel rates at 5′ and 3′ Junctions after 1,010 bp fragment KI using different donors by TIDE measurement at AAVS1 locus among HEK293T cells, HepG2 cells, K562 cells, and Jurkat cells. (B) High throughput amplicon-seq analyses for the on-target editing events as determined using dsDNA, odsDNA and ssDNA donors. The amplicon-seq library was constructed with specifically designed PCR primers spanning the junctions. (C) Schematic diagram showing the Genome-wide insertion site sequencing (GIS-seq) library construction pipeline for genome-wide off-target detection. The individual 1,010 bp dsDNA or 1,010 bp odsDNA donor was transduced into HEK293T cells through nucleofection. Following the sonication of the genomic DNA, the library was constructed with a specific primer (forward or reverse) against the donor and an adaptor primer. (D) Representative GIS-seq result showing plus/minus read distribution at on-target locus. (E) GIS-seq revealed that odsDNA donors had similar or even lower off-target integration at Lamin A/C locus than dsDNA donors in two biological experiments. Data and error bars in A and B indicate the mean and SD of three independent biological replicates.