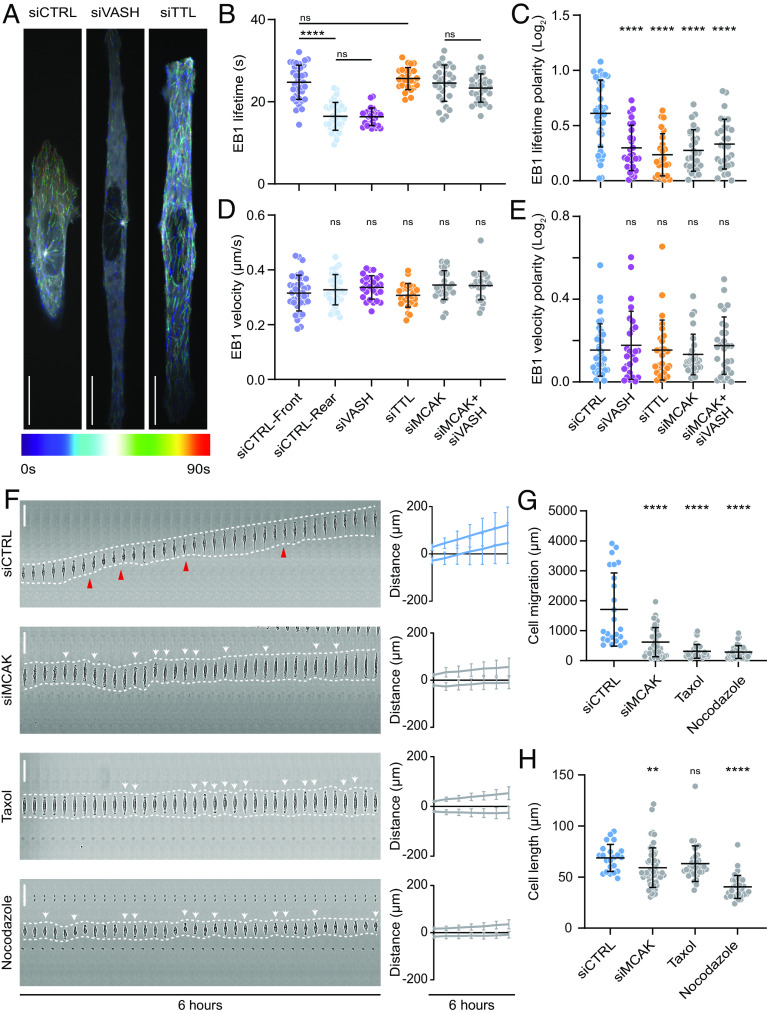
Fig. 3.
Microtubule detyrosination regulates microtubule dynamics during directed cell migration. (A) Representative color-coded temporal projections of U2OS EB1-GFP cells on linear micropatterns treated with indicated siRNAs. (Scale bar, 20 µm.) (B, C) Quantification of microtubule stability (EB1 lifetime) (B) and its polarity (C) by manual tracking of EB1-GFP comets in U2OS EB1-GFP cells on linear micropatterns subjected to indicated siRNA treatments. (N, n): siCTRL (38,4), siVASH (26,3), siTTL (25,3), siMCAK (29,3), siMCAK + siVASH (28,3). (D, E) Quantification of EB1-GFP comet velocity (D) and its polarity (E) in U2OS EB1-GFP cells on linear micropatterns treated following indicated treatments. (N, n): siCTRL (38,4), siVASH (26,3), siTTL (25,3), siMCAK (29,3), siMCAK + siVASH (28,3). (F) Representative kymograph of RPE-1 cell migration on 5-µm linear micropatterns following treatments with indicated conditions. White dotted lines follow the edges of the cell, red arrow heads mark retraction events at the cell rear, and white arrows indicate change in migration direction. The combined kymograph trajectories with mean ± SD are shown on the Right. (Scale bar, 50 µm.) (N, n): siCTRL (133,18), siMCAK (40,3), Taxol (35,3), and nocodazole (36,3). (G and H) Scatter plot of total cell migration distance (G) and cell length (H) of RPE-1 cells with indicated treatments after 6 h on 5-µm linear micropatterns. (N, n): Cell migration—siCTRL (24,3), siMCAK (40,3), Taxol (35,3), and nocodazole (36,3); cell length—siCTRL (22,3), siMCAK (55,4), Taxol (35,3), and nocodazole (36,3). N, number of cells; n, number of independent experiments. The scatter plots include mean ± SD. ns, not significant, **P ≤ 0.01 and ****P ≤ 0.0001.