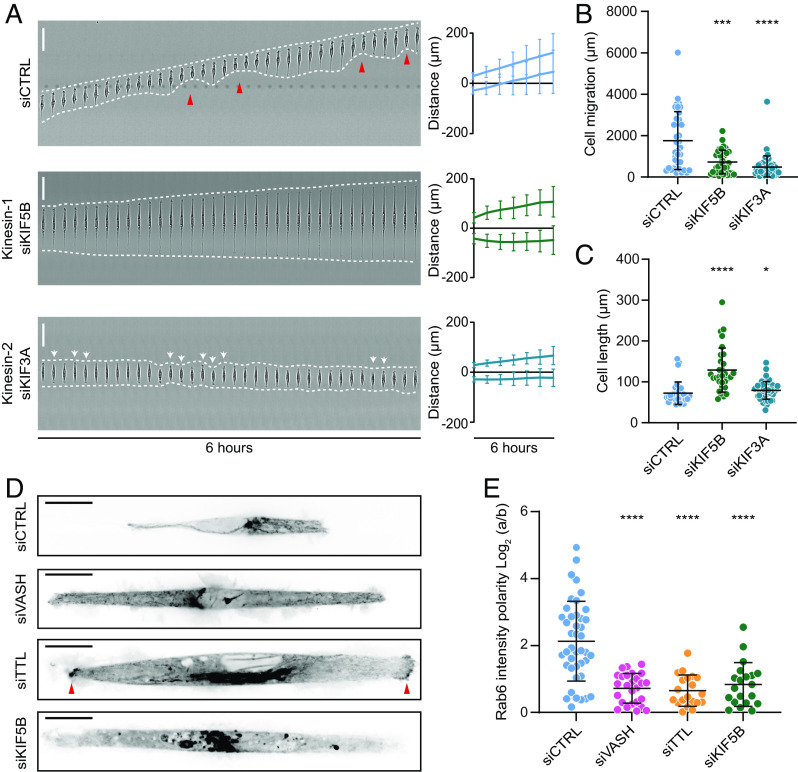
Fig. 4.
Microtubule detyrosination regulates kinesin-1-based intracellular transport. (A) Representative kymograph of RPE-1 cell migration on 5-µm linear micropatterns following treatments with control and kinesin-specific siRNAs. White dotted lines follow the edges of the cell, and red arrow heads mark retraction events at the cell rear. The combined kymograph trajectories with mean ± SD are shown on the Right. (Scale bar, 50 µm.) (N, n): siCTRL (133,18), siKIF5B (33,3), and siKIF3A (49,3). (B and C) Scatter plot of total cell migration distance (B) and cell length (C) of RPE-1 cells with indicated treatments after 6 h on 5-µm linear micropatterns. (N, n): Cell migration—siCTRL (36,4), siKIF5B (33,4), and siKIF3A (49,4); cell length—siCTRL (31,4), siKIF5B (31,4), and siKIF3A (49,4). (D) Representative temporal projections of GFP-Rab6 transfected RPE-1 cells treated with indicated siRNAs on linear micropatterns. The red arrowhead highlights accumulation of GFP-Rab6 at the cell edges. (Scale bar, 20 µm.) (E) Quantification of polarity of GFP-Rab6 intensity at the cell edges (a/b) in RPE-1 cells subjected to indicated siRNA treatments. (N, n): siCTRL (42,5), siVASH (26,4), siTTL (20,3), and siKIF5B (20,3). N, number of cells; n, number of independent experiments. The scatter plots include mean ± SD. *P ≤ 0.05, ***P ≤ 0.001, and ****P ≤ 0.0001.