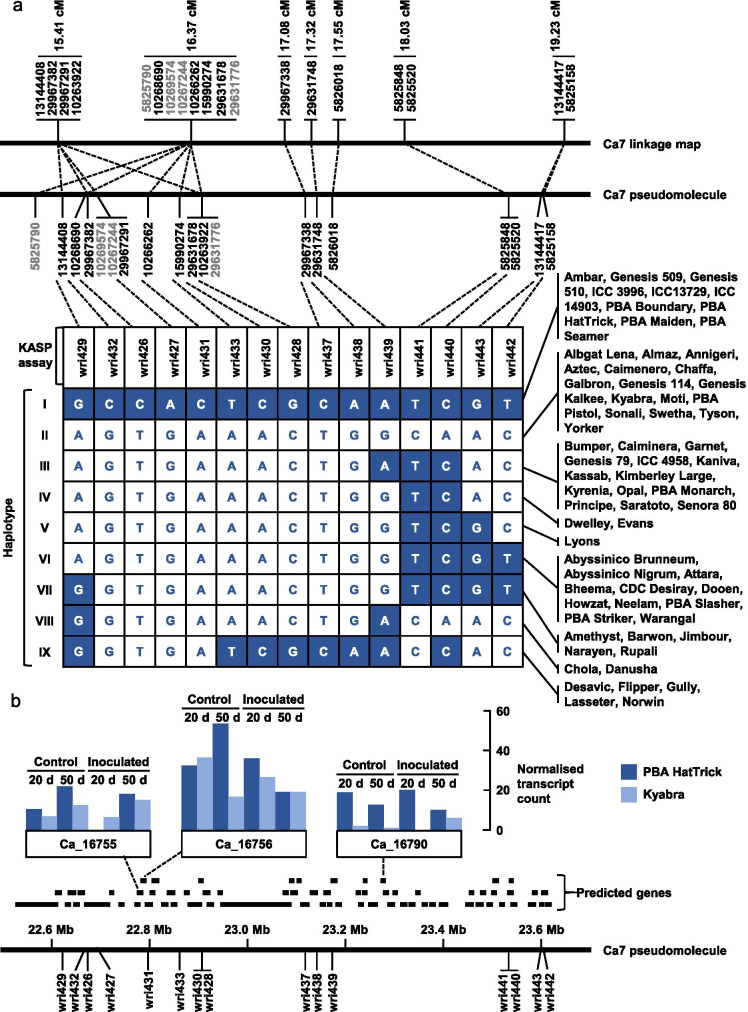
Fig. 4.
a Genetic positions of 19 genotyping-by-sequencing markers that are within 2 cM of the estimated position of a QTL for resistance of chickpea to Pratylenchus thornei, physical positions at which those markers were anchored on the Ca7 pseudomolecule of version 2.6.3 of a genome assembly for kabuli-type chickpea; and haplotypes observed among 68 chickpea accessions genotyped with KASP assays for 15 of those markers. In the haplotypes, nucleotides shown in white text on a dark background are the same as those carried by the partially resistant parent PBA HatTrick and nucleotides shown in dark text on a white background are the same as those carried by the susceptible parent Kyabra. b Physical positions of 69 genes predicted with high confidence within a 1.03 Mb region of the Ca7 pseudomolecule, with transcript abundance data (from Channale et al. 2021) for three of those genes