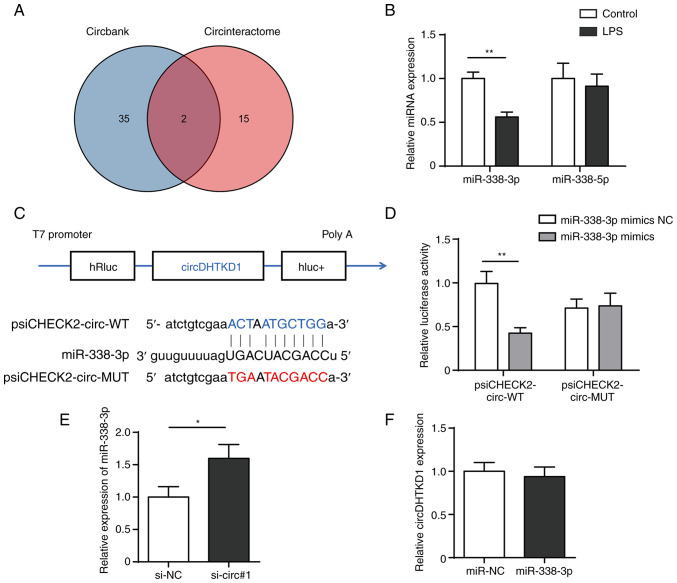
Figure 4.
circDHTKD1 serves as a sponge for miR-338-3p. (A) CircBank and CircInteractome databases were used to screen miRNAs that potentially interact with circDHTKD1. (B) RT-qPCR analysis of expression of miR-338-3p and miR-338-5p in LPS-treated BEAS-2B cells. (C) Schematic of predicted binding sites of circDHTKD1 and miR-338-3p. (D) Dual-luciferase reporter assay was used to validate the interaction between circDHTKD1 and miR-338-3p. (E) miR-338-3p and (F) circDHTKD1 levels were detected using RT-qPCR. *P<0.05, **P<0.01. RT-qPCR, reverse transcription-quantitative PCR; hRluc, Renilla luciferase reporter gene; WT, wild-type; MUT, mutant; NC, negative control; si, small interfering; DHTKD1, dehydrogenase E1; circ, circular; miR, microRNA; LPS, lipopolysaccharide.