The authors show T cells expressing a modified IgG receptor and a signaling coreceptor efficiently mediate potent antitumor immunity without on-target, off-tumor activity, suggesting a way to address challenges of CAR T-cell therapy for solid tumors.
Abstract
The pioneering design of chimeric antigen receptor (CAR) T-cell therapy demonstrated the potential of reprogramming the immune system. Nonetheless, T-cell exhaustion, toxicity, and suppressive microenvironments limit their efficacy in solid tumors. We previously characterized a subset of tumor-infiltrating CD4+ T cells expressing the FcγRI receptor. Herein, we detail engineering of a receptor, based on the FcγRI structure, allowing T cells to target tumor cells using antibody intermediates. These T cells showed effective and specific cytotoxicity only when an appropriate antibody was added. Only target-bound antibodies activated these cells, while free antibodies were internalized without activation. Their cytotoxic activity was correlated to target protein density, therefore targeting tumor cells with high antigen density while sparing normal cells with low or no expression. This activation mechanism prevented premature exhaustion. Furthermore, during antibody-dependent cytotoxicity these cells secreted attenuated cytokine levels compared with CAR T cells, thereby enhancing their safety profile. These cells eradicated established melanomas, infiltrated the tumor microenvironment, and facilitated host immune cell recruitment in immunocompetent mice. In NOD/SCID gamma mice the cells infiltrate, persist, and eradicate tumors. As opposed to CAR T-cell therapies, which require changing the receptor across different types of cancer, our engineered T cells remain the same across tumor types, while only the injected antibody changes. Overall, we generated a highly flexible T-cell therapy capable of binding a wide range of tumor cells with high affinity, while preserving the cytotoxic specificity only to cells expressing high density of tumor-associated antigens and using a single manufacturing process.
Introduction
The importance of T cells in the elimination of hematologic cancers has been demonstrated in clinical trials, where a higher incidence of disease relapse was observed in T cell–depleted transplants compared with T cell–replete recipients (1–3). Pioneering therapies based on the de novo expansion of tumor-infiltrating cytotoxic T cells and their reinfusion into patients in combination with high-dose IL2 demonstrates the capability of the immune system to mediate tumor rejection (4, 5). However, this is a harsh and complicated procedure, has only been partially successful, and is restricted to the expansion of T-cell clones that the host immune system can spontaneously activate to infiltrate into the tumor sites. Moreover, injected T cells must be expanded ex vivo into billions—a process that lasts between 3 to 5 weeks, often too long for patients (6). Most tumors express mutated tumor-associated antigens (TAA) that can be targeted by T cells (7–9). Nonetheless, attempts to harnesses and augment immune mechanisms to fight malignancies using expansion of T cells that are specific for TAAs on blood cancers have been limited to a subset of responding individuals, in only a small number of malignancies (10, 11). One of the reasons is that this therapy depends on the limited host repertoire of naturally occurring T-cell receptors (TCR) against TAAs and their inherently low affinity (typically ranging at 10−4–10−6 mol/L; ref. 12). To overcome these limitations, pioneering work has demonstrated that T cells can be engineered to express chimeric antigen receptors (CAR) that recognize tumor antigen at affinities comparable with that of antibodies, ranging up to approximately 10−9 mol/L (13). This has resulted in remarkable clinical responses observed in patients treated with CAR engineered T cells, establishing this cell therapy as one of the most promising clinical approaches for cancer (14–16).
CARs are modular proteins composed of three functional domains: an antigen-binding domain that results from linking the single variable fragments of the light and heavy chains of an antibody (ScFv), a transmembrane domain, and one or more signal transduction domains (13). The first-generation of CARs used CD3ζ, or Fcγ receptor γ-chains as a single signal transduction domain. While effective in preclinical studies, clinical efficacy was greatly improved upon incorporation of co-stimulatory domains in second-generation CARs (CD28, 4–1BB) and additional signaling domains in third-generation CARs (CD27, ICOS; refs. 13, 17). Nonetheless, therapies using CAR engineered T cells are currently effective only with hematologic malignancies, and about half the patients suffer a relapse within a few months (16, 18). Importantly, after relapse following anti-CD19 CAR T-cell therapy for lymphoma, the cancer cells typically either lose or alter the expression levels of the CD19 antigen as an escape mechanism (19–21). Additional limitations stem from the lack of sufficient identified tumor-specific antigens, as well as from constitutive tonic signaling in the absence of ligands; these limitations significantly reducing the clinical use of CAR T-cell therapy, while also resulting in off-site toxicity and T-cell exhaustion (17, 22). While the successes of CAR T-cell therapy highlight the clinical benefits of directing T cells against cancer, an urgent need remains to develop treatments capable of eradicating additional tumors, specifically solid tumors, that feature a higher safety profile and do not rely exclusively on the host T-cell repertoire.
Recently, we discovered a subset of CD4+ T cells that directly kill tumor cells coated with antibodies (23). These cells express the high-affinity Fcγ receptor (FcγRI), which enables them to bind antibody-coated tumor cells and secrete lytic granules resulting in tumor lysis. These cells, however, could not be expanded using T-cell mitogens and a different strategy is required to use them in immunotherapy. Here, we developed an altered version of FcγRI that endows peripheral blood T cells with remarkable antibody-dependent cell cytotoxic (ADCC) abilities against cells coated with antibodies. Moreover, ADCC killing was restricted to tumor cells expressing a high antigen density, while leaving cells expressing physiologic or low antigen density intact. Administration of these cells in combination with tumor-binding antibodies, into immunocompetent mice bearing established tumors, induced tumor eradication and long-lasting immunity by facilitating the recruitment of host immune cells, providing a demonstration of the potency of this strategy. This technology has the potential to treat a wide range of cancers as well as to target refractory cancers that have lost tumor antigens by switching to alternative, clinically approved antigen-targeting antibodies such as cetuximab and trastuzumab, while providing a better safety profile and a higher potency compared with CAR T-cell therapies.
Materials and Methods
Mice
Wild-type (WT) C57BL/6 mice were obtained from Envigo (Jerusalem, Israel), and from Jackson Laboratories (Bar Harbor, ME, USA). NOD/SCID gamma (NSG) mice were obtained from Jackson Laboratories. All mice were housed in an American Association for the Accreditation of Laboratory Animal Care–accredited animal facility and maintained in specific pathogen-free conditions. Male and female 8-to-12-weeks-old mice were used in all experiments. Animal experiments using (WT) C57BL/6 mice were approved by the Tel Aviv University. Animal experiments using NSG mice were approved by the Israeli national council for experiments on animal subjects.
Cell lines
B16F10 cells (purchased in 2017, ATCC), HEK-293FT (purchased in 2017, Thermo Fisher Scientific, Waltham, MA) and HEK GP2 (purchased in 2021, Takara) were cultured in DMEM (41965–039, GIBCO). HT29 (purchased in 2020, human colon adenocarcinoma HTB-38, ATCC) and SK-BR3 (purchased in 2021, ATCC) were cultured in McCoy's 5A (01–075–1A, Sartorius). A549 were cultured in F-12K (01–095–1A, Sartorius). H1299, NCI-N87, HeLa and HCC1954 were purchased from the ATCC (2021) and cultured in RPMI1640 (21875–034, GIBCO). A431 and JIMT-1 were purchased from AddexBio (2021) and cultured in DMEM. All cell lines were used in passages 2–10. All the above media were supplemented with 10% heat-inactivated FBS (12657–029 GIBCO), 2 mmol/L l-glutamine (25030–024 GIBCO), and 100 μg/mL penicillin/streptomycin (15140–122, GIBCO). Cells were routinely tested for mycoplasma using EZ-PCR Mycoplasma Test Kit (20–700–20 Biological Industries, Israel) according to the manufacturer's instructions. HT29 cells were transduced by lentivirus containing pLVX-H2B-tdTomato and/or pLVX-HER2 (extracellular + transmembrane + 7 residues of intracellular domain) and were sorted by FACS (BD FACSAria III, BD Biosciences, Franklin Lakes, NJ) for the high-expressing population. All human cell lines were analyzed for authentication (Technion Genomic center, Technion, Haifa Israel), using the Promega GenePrint 24 System in order to determine short tandem repeat profile of 23 loci plus Amelogenin for gender determination (X or XY). In addition, the male-specific DYS391 locus was included to identify null Y allele results for Amelogenin. DNA sample from the kit (2800M Control DNA) was included in the analysis and served as positive control for the PCR step. No DNA template was also included as negative control. The results were analyzed using the 3500xl Genetic Analyzer v 1.0 (Life Technologies) and GeneMapper IDX software v 1.3 (Life technologies). Allelic ladder was included in the run. Results were compared with the Cellosaurus, a Cell-Line Knowledge Resource (Bairoch A., J. Biomol Tech, 2018) and are summarized in Supplementary Table S1. Mouse cells (B16F10) were not authenticated in-house but by the manufacturer (ATCC) and used at early passage (P < 5).
Lentiviral infection
HEK-293FT cells were transfected with pLVX plasmids containing H2B-tdTomato or HER2 under EF1a promoter together with psPAX2 (Addgene, plasmid # 12260) and pCMV-VSV-G (Addgene, plasmid # 8454). Media-containing viruses were collected following 24 and 48 hours. For transduction of HT29, cells were incubated with viruses and 100 μg/mL polybrene (107689, Sigma Aldrich, Merck, Israel) for 30 minutes followed by 30 minutes centrifugation at 400 × g before medium was replaced. Following 3 days, cells that expressed HER2/tdTomato were sorted by FACSAriaII using anti-HER2 (clone 24D2) conjugated to BV421 (324420, BioLegend).
Constructs design
Mouse sequences encoding FcγRI-extracellular-transmembrane-FcRγ−intracellular-T2A-FcRγ (AG2G) full coding sequence (CDS) all in frame were synthesized by GeneART (Thermo Fisher Scientific) into pMK vectors and were further cloned into pMIGII using EcoRI/XhoI sites. Human sequences encoding: (A2G) FcγRI-extracellular-transmembrane-intracellular-T2A-FcRγ-extracellular-transmembrane-intracellular CDS in frame, (AG2G) FcγRI-extracellular-transmembrane-FcRγ-intracellular-T2A-FcRγ-extracellular-transmembrane-intracellular CDS all in frame, (AGO2GO) FcγRI-extracellular-transmembrane-FcRγ-intracellular-OX40-intracellular-T2A- FcRγ-extracellular-transmembrane-intracellular–OX40-intracellular CDS all in frame, (CD28) FcγRI-extracellular-transmembrane-FcRγ- intracellular-CD28-intracellular-T2A-FcRγ-extracellular-transmembrane-intracellular–CD28-intracellular CDS all in frame, (4–1BB) FcγRI-extracellular-transmembrane-FcRγ-intracellular-4–1BB-intracellular-T2A-FcRγ-extracellular-transmembrane-intracellular–4–1BB- intracellular CDS all in frame, and anti-HER2 CAR was contracted by using the trastuzumab derived scFv (variable light chain-G4S x3 linker variable heavy chain)-CD8a-linker-transmembrane-CD28-intracellular-4–1BB-intracellular-CD3ζ-intracellular, CD16(158V)-extracellular-CD28-transmembrane-CD3ζ-intracellular (ACTR707) were synthesized by GeneART (Thermo Fisher Scientific) into pMK vectors and were further cloned into pMIGII using EcoRI/XhoI sites upstream to IRES-GFP sequences or into pMSVG.1 vector cut with NcoI/NotI. Sequences were verified by sequencing (HyLabs Israel). Histone H2B sequence was amplified with AATAACACTAGTGCCACCATGCCTGAACCGGCAAAAT and AACAACCCCGGGACTTGTCGTCATCGTCTTTGT primers and cloned into pLVX vector (Clontech) containing EF1 promoter into SpeI/XmaI sites in frame with tdTomato. Human extracellular domain and transmembrane domain plus 7 residues of intracellular of HER2 were synthesized by GeneART (Thermo Fisher Scientific) into pMK vectors and cloned into pLVX vector (Clontech) containing EF1 promoter into SpeI/NotI sites. Sequence was verified by MSC\v forward and tdTomato reverse primers (HyLabs Israel). Construct sequences can be found in Patentscope under the following accession numbers: WO/2020/188570 and WO/2021/260696.
Retroviral infection
Mouse total CD3+ T cells were isolated from mouse spleen using magnetic beads (BioLegend, #480099) according to the manufacturer's protocol and transduced with the above constructs as follows: HEK293FT cells were plated on 10-cm culture plates and co-transfected with 2:1 molar ratio of pMIGII and PCL-Eco plasmids using Polyplus jetPRIME reagent (Polyplus transfections, #1). After 24 hours, media was replaced with DMEM supplemented 10% FBS and 0.075% sodium bicarbonate. Media-containing viruses were collected after 24 hours and 48 hours and centrifuged for 1 hour at 100,000 × g. Pellet was resuspended gently in 1 mL media and let to recover overnight at 4°C. Prior to infection, splenic CD3+ T cells were incubated on plates precoated with anti-CD3 (BioLegend, clone 17A2, #100238; 0.5 μg/mL) in T-cell medium, which is composed of RPMI supplemented with 10% FBS, 0.075% sodium pyruvate (Thermo Fisher Scientific, # 11360039), 1% nonessential amino acids (Gibco, # 11140–035), 1% insulin-transferrin-selenium (Gibco, #41400–045), and 50 μmol/L beta-mercaptoethanol (Thermo Fisher Scientific, #31350010), containing IL2 (Peprotech, # 200–02–250, 1,000 IU/mL). Next, 0.3 mL of concentrated retroviruses was added to every 2×106 T cells with 10 μg/mL polybrene. Cells were incubated for 30 minutes in 37°C, 5% CO2 and centrifuged at 37°C 1,200 rpm for 1 hour. Next, 80% of medium was replaced and T cells were cultured for an additional three days in T-cell medium containing IL2 (1,000 IU/mL). For infection of human peripheral blood mononuclear cell (PBMC), 7×106 GP2 cells were cultured in PLL-coated 10-cm plates, transfected using Lipofectamine 2000 (Thermo Scientific) with 9-μg pMIGII (backbone originated from plasmid received as a gift from Dario Vignali, University of Pittsburgh) or pMSGV.1 plasmid (received from Nicholas P. Restifo, the NCI, NIH, Bethesda) together with 4-μg FBSalf-RD114 envelope plasmid (received from Nicholas P. Restifo). Forty-eight hours and 72 hours later, supernatants were collected and concentrated using amicon ultra 100kDa MWCO (Millipore), and 2×105 cells were infected in 24-well suspension plates coated with Retronectin (Takara, #T100B) for 24 hours, transferred to 12-well culture plates in T-cell medium with 300 U/mL IL2.
Primary human cells
Human PBMCs were purchased from STEMCELL Technologies and CellGeneration, activated for 48 hours with 50 ng/mL Ultra-LEAF Purified anti-human CD3 (OKT3, BioLegend), together with 300U IL2, followed by retrovirus transduction. Human Primary Alveolar Epithelial Cells (Catalog no. H-6053) and Human Primary Kidney Epithelial Cells (Catalog no. H-6034) were obtained from CellBiologics, (Chicago, IL) and cultured in Complete Human Epithelial Cell Medium (H6621 CellBiologics, Chicago, IL) containing supplements and 100 U/mL penicillin, and 100 μg/mL streptomycin on gelatin-coated cell culture flasks (0.2% solution on Greiner Bio-One flasks). The cells were incubated at 37°C until they reached 90% confluency, and were used from passages 4–10. Human Pulmonary Microvascular Endothelial cells (HPMEC, catalog no. 3000), Human Renal Glomerular Microvascular Endothelial cells (HRGEC, catalog no. 4000) and Human Cardiac Microvascular Endothelial cells (HCMEC, catalog no. 6000), were obtained from ScienCell, (Provitro AG, Berlin, Germany) and cultured in ECM (ScienCell, Provitro AG, Berlin, Germany, #1001) containing supplements, and 100 U/mL penicillin and 100 μg/mL streptomycin on fibronectin-coated cell culture flasks (Greiner Bio-One flasks). CD4 and CD8 cells were separated from the PBMCs by Mojosort negative magnetic selection kits according to the manufacturer's instructions (BioLegend #480010, #480012). For γδ T-cell preparation, PBMCs were activated with zoledronate (Taro, lot:HAC2249A) for 5 days followed by retroviral transduction with pMSGV1-AG2G vector. On day 9, cells were isolated using human γδ T cell isolation kit (StemCell EasySep #19255).
Confocal microscopy
Human PBMCs were stained with anti-human CD3 (HIT3a, BioLegend), allophycocyanin–anti-human CD64 (10.1, BioLegend), and Alexa Fluor 647 anti-human CD16 (3G8, BioLegend). HT-29 were stained with Alexa Fluor 488 anti-human CD340 (erbB2/HER-2, 24D2, BioLegend), CAR T cells were stained by Human HER2/ErbB2 Protein (ECD) Biotinylated (Sino Biological, #10004-HCCH-B) and Alexa Fluor 594 Streptavidin (BioLegend, #405240). Images were collected using a Zeiss LSM800 confocal laser scanning microscope using Objective Plan-Apochromat 20x/0.8 DIC M27 and solid-state laser lines, and analyzed using ZEN software (Carl Zeiss Microscopy, version ZEN 2.6 blue).
Analysis of human T-cell cytotoxicity and activation
IncuCyte imager killing assays were conducted by culturing 104 HT-29 H2B-tdTomato for cell count analysis or nonlabeled HT-29 cells for confluency analysis in 96-well plates; 24 hours later human αβ T cells or γδ T cells were added in the specified ratios, with or without the specified concentration of trastuzumab (Roche), cetuximab (Merck), avelumab (Merck Serono), sofituzumab (Creative Biolabs, #TAB-H63), necitumumab (Creative Biolabs TAB-119), or rituximab (Genentech). Cells were imaged by incuCyte S3 imager (Sartorius), and images were then used to calculate numbers of target cells or confluency by incuCyte software (version 2021A, basic analyzer). Human T-cell activity was also evaluated by measuring cytokine levels using the following ELISA kits according to the manufacturer's instructions: Human TNF-alpha DuoSet ELISA (DY210, R&D Systems), Human IL2 DuoSet (DY202, R&D Systems), Human IFN-gamma DuoSet ELISA (DY285, R&D Systems), Human Perforin ELISA flex (3465–1H, Mabtech) and Human Granzyme B DuoSet ELISA kit (R&D systems, DY2906). IVIG (Intratect 50 gr/L, lot:B791319P01) was added at increasing concentrations to the killing assay coculture. Supernatants from killing assay cocultures were collected for analysis of cytokine levels, alternatively, cytokine levels were determined by multiplex bead analysis using Human Luminex Discovery Assay (12 PLEX; R&D Systems, LXSAHM-12) and analyzed by MAGPIX 4.2. The following cytokines and chemokines were included: CXCL10, Granzyme B, IL2, IL4, IL5, IL10, IL21, GM-CSF, IFNγ, IL6, IL17, TNFα. For activation of effector cells on plastic-bound ligands, monoclonal antibodies or recombinant Human HER2/ErbB2 Protein (ECD) Biotinylated (Sino Biological, #10004-HCCH-B) were incubated on Maxisorp ELISA 96-well plates (Nunc Thermos Fisher Scientific) in PBS for 24 hours, washed three times and 105 T cells were incubated for 24 hours in 150 μL RPMI medium supplemented with 10% FBS.
Flow cytometry
Mouse isolated T cells were analyzed using flow cytometry (CytoFLEX, Beckman Coulter, Lakeview Indianapolis, IA) for expression of GFP following pMIGII viral transduction or by staining with Alexa Fluor 647 anti-mouse CD64 (FcγRI) (X54–5/7.1, BioLegend), Brilliant Violet 421 or APC or PE anti-human CD64 (10.1, BioLegend), FITC-anti-human CD3 (UCH1, BioLegend), Brilliant Violet 421-anti-CD4 (RPA-T4), APC–anti-CD8 (RPA-T8), PE-anti-γδ-TCR (BioLegend, #331209), anti–PD-1 (BioLegend, #329924), anti–LAG-3 (BioLegend, #369208), anti-Tim3 (BioLegend, 345016). Intracellular staining of FcRγ was performed using Milli mark anti-FcεRI γ subunit -FITC (Merck, FCABS400F). Datasets were analyzed using FlowJo software (Tree Star, version 10.7.2).
Absolute quantification of cell-surface antigens
5 × 105 cells were stained for HER2 membrane protein using anti-HER2 (324401, BioLegend) according to the manufacturer's instructions for use. Mouse IgG1 (302301, BioLegend) was used as a negative control antibody. QIFIKIT (Dako, Denmark) was used to determine the HER2 expression of cell lines according to the manufacturer's instructions. Beads and cells are stained with secondary FITC labeled antibody. Flow cytometry analysis was performed on a CytoFLEX cytometer (Beckman Coulter, Lakeview Indianapolis, IA). FITC fluorescence intensity of labeled cells was determined on a FL1 detector (530/30 band filter). Mean fluorescence intensity (MFI) is calculated by FlowJo software and correlated with the antibody binding capacity (ABC) of the beads. The MFI of each bead group was used to plot a calibration graph of MFI against ABC of each bead by linear regression using MS Excel v16.7 (Microsoft). Unknown sample antigen quantities were calculated by extrapolation from the calibration curve.
Immune competent melanoma efficacy model
2×105 B16F10 cells suspended in 50 μL DMEM were injected subcutaneously into C57BL/6 mice above the right flank and the size of growing tumors was measured twice a week using calipers. When tumors reached 120 mm2, mice were sacrificed for ethical considerations. Animal were injected intravenously with 106 uninfected (sham) or retrovirally infected with pMIGII-alpha-gamma-2A-Gamma CD3+ T cells, with or without intraperitoneal injection of 300 μg anti–TRP-1 antibody (clone TA99). Treatment was applied at days 7, 11, and 14 post tumor injection.
Immune competent melanoma infiltrate analysis model
C57BL/6 male mice were injected subcutaneously with 2×105 B16F10 cells and tumors were allowed to grow for 6 days until they reached approximately 25 mm2 in diameter. Four groups (n = 6 each) were injected with: untreated (saline intraperitoneally and intravenously), sham+TA99 (non-transduced T cells intravenously + TA99 300 μg per mouse intraperitoneally), AG2G (transduced T cells intravenously), AG2G+TA99 (transduced T cells intravenously + TA99 300 μg per mouse intraperitoneally). The AG2G transduced T-cell administration was 2×106 CD3 splenocytes from CD45.1 mice (kind gift from Adi Barzel, Tel Aviv University) infected with the AG2G construct (engineered cells were approximately 20% of total injected T cells). On day 3 and day 7 following T-cell injection, tumors were extracted and enzymatically digested by 2% of collagenase (MERK, # C5138) to obtain a single-cell suspension. Cells were then washed in PBS and mixed with 20% Percoll gradient to reduce dead cell and cell debris.
Immune-deficient xenograft efficacy model
2.5×106 NCI-N87 gastric carcinoma cells were injected subcutaneously into NSG mice above the right flank and the size of growing tumors was measured three times a week using calipers. Mice were treated when tumors reached a size of 70 to 120 mm3 (n = 6/group). Human AG2G cells used for treating the mice were produced from PBMC as described in “Retroviral infection” and in Supplementary Fig. S1. Three treatments were administered in the following groups: control (saline injection intraperitoneally and intravenously), trastuzumab – (250 μg/mouse/dose intraperitoneally), trastuzumab+AG2G (250 μg/mouse/dose intraperitoneally + 5×106 AG2G cells/mouse/dose intravenously). Terminal bleedings were collected at day 42 and analyzed: Chemistry - cobas 6000 analyzer (Roche) and CBC - Advia (Siemens).
Immune-deficient xenograft model for persistence analysis
2.5×106 NCI-N87 gastric carcinoma cells were injected subcutaneously into NSG mice above the right flank and the size of growing tumors was measured 3 times a week using calipers. Mice were treated with a single administration when tumors reached a size of 150 to 200 mm3 (n = 12/group). There were two treatment groups: AG2G (5×106 AG2G cells/mouse intravenously) and trastuzumab+AG2G (250 μg/mouse intraperitoneally + 5×106 AG2G cells/mouse/dose intravenously). On days 1, 7, 14, and 28, 3 mice from each group were sacrificed and blood as well as tumor, spleen, liver, kidney, lung, brain, and heart were harvested and RNA was extracted by RNA nucleoSpin (Macherey Nagel, # MAN-740955.250). RNA was reverse transcriberd to cDNA using High Capacity RNA-to-cDNA Kit (Applied Biosystems, # 4387406). cDNA was used as template for qPCR as described herein.
Histochemistry and IHC
Tumors and healthy organs from NSG tumor-bearing mice were paraffin embedded and sectioned at 5-μm thick. The sections were stained with hematoxylin and eosin (H&E) and studied using Olympus BX51 microscope equipped with the 12MP SPOT InSight CMOS camera. Necrosis was evaluated in H&E-stained sections according to the necrotic area in relation to the total tumor area: Grade 0, 0% to 10%; Grade 1, 10% to 25%; Grade 2, 25% to 50%; Grade 3, 50% to 75%; Grade 4, >75%. HER2 staining was performed using mouse monoclonal anti-human HER2 (Cell Signaling Technology, catalog no. 76799). HER2 was evaluated in HER2 stained sections according to the following scale: IHC 3+ (strong positive): tumor displays complete, intense circumferential membranous staining in >10% of tumor cells, IHC 2+: (weak to moderate) complete membrane staining observed in > 10% of invasive tumor cells, IHC 1+: incomplete faint membrane staining and within >10% of invasive tumor cells, IHC 0: no staining observed or incomplete faint / barely perceptible membrane staining within ≤10% of invasive tumor cells. CD3 staining was performed using Rabbit monoclonal antibody (EP449E) to human CD3 epsilon (Abcam, catalog no. ab25959,). heart, kidney, liver, spleen and lung from C57BL/6 mice were paraffin embedded and sectioned at 5-μm thick. The sections were stained with H&E and studied using slide scanner Aperio Versa 200 (Leica Biosystems, model DM6000 B).
qPCR
qPCR amplification and analysis were performed using the StepOne Real-Time PCR Systems (Thermo Fisher, USA). All reactions (20-ng cDNA template and three replicates) were performed using PowerTrack SYBR green master mix (Thermo Fisher, USA, #A46109) using the StepOne Real-Time PCR Systems (Thermo Fisher, USA) according to the manufacturer's instructions. The PCR reaction conditions were as follows: preincubation at 95°C for 10 minutes; 40 cycles at 94°C for 10 seconds, 60°C for 30 seconds, and 72°C for 30 seconds and a final extension at 72°C for 3 minutes. Fluorescence was measured at the end of each annealing step. Amplification was followed by a melting curve analysis with continuous fluorescence data acquisition during the 56° to 80°C melt. The raw data were analyzed with the StepOne Real-Time PCR Systems (Thermo Fisher, USA), and the gene expression levels were normalized to mouse beta actin using the 2−ΔΔCt method. The following primers used: AG2G CD8-alpha F: CCTTACCAGTGACCGCCTTG; AG2G CD8-alpha R: CTGAGCAATGGTAGGTGCCA (Amplifies only AG2G DNA and RNA but not CD64 DNA or RNA). Mouse actin cDNA F: GGCTGTATTCCCCTCCATCG, Mouse actin cDNA R: CCAGTTGGTAACAATGCCATGT.
CyTOF
Purified antibodies were obtained from commercial vendors and labeled with metal isotopes using the MAXPAR X8 chelating polymer kit (Fluidigm Corporation) following the manufacturer's instructions. A full list of antibodies, their elemental isotope tags, clone, and source are listed in Supplementary Table S2. Tumor samples were obtained from mice as described in “Immune competent melanoma infiltrate analysis model” section. Approximately 3 million cells per sample were used for staining and incubated with isotope-labeled antibodies. For viability staining, cells were washed with CSM Cell Staining Buffer (Standard Biotools, 201068) and incubated for 15 minutes in 500 μL of 250 nmol/L Rhodium solution (Standard Biotools, 201103A) at room temperature, following washing. The antibodies to surface proteins were added in a total reaction volume of 100 μL; the cells were incubated at room temperature for 30 minutes, washed and permeabilized with Transcription Factor Buffer Set (BD Bioscience, 562574) for 45 minutes at 4°C. After permeabilization, the cells were incubated with antibodies to the intracellular proteins for 45 minutes on ice. The washed cells were then fixed in 1.6% PFA for 10 minutes, washed and resuspended in 125 nmol/L Ir DNA intercalator (Standard Biotools, 201192A), incubated overnight at 4°C. Prior CyTOF acquisition, samples were washed once in CSM, and twice more in Cell Acquisition Solution (Standard Biotools, catalog no. 201240). Samples were then passed through a cell strainer and spiked with internal metal isotope normalization beads EQ Four Element Calibration Beads (Standard Biotools, 201078) following acquisition on a Helios mass cytometer (Fluidigm Corporation). Acquired data was signal normalized and concatenated using the Helios Software (Standard Biotools, CyTOF Software version 7.0) and uploaded into Cytobank (Cytobank Inc.) for further QC data processing. Gaussian parameters of the Helios system were used for doublet exclusion, Ir+ and Rh- were used for gating out of dead cells and normalization beads. Data was transformed using an arcsinh(X/5) transformation. Single live intact cells were then used for data analysis.
Statistical analyses
All statistical analyses and graphs were performed by GraphPad Prism 9. Each experimental group consisted of at least 3 mice. Significance of results was determined using the nonparametric one-way ANOVA with Dunnet and Holm-Sidak correction, when multiple groups are analyzed, two-way ANOVA with Tukey's and Sidak correction for multiple comparisons.
Data availability
The data generated in this study are available within the article and its Supplementary Data files or upon request from the corresponding author.
Results
An altered version of FcγRI endows peripheral blood T cells with superior ADCC capacities
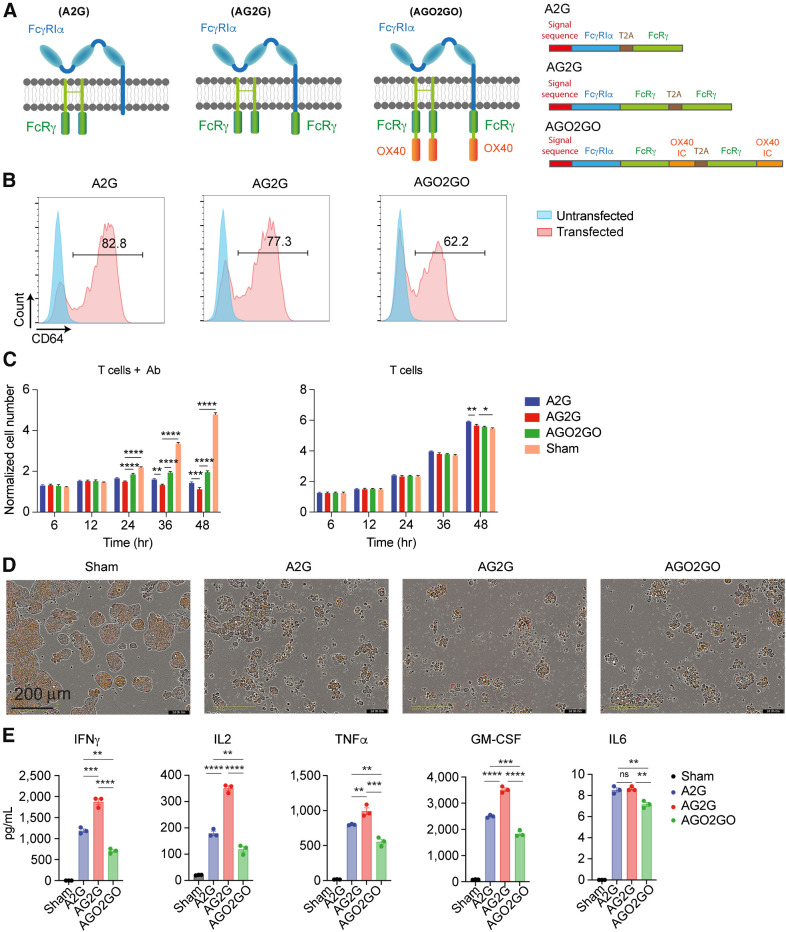
We have previously described a subpopulation of T cells in both human and mouse tumors that express the high-affinity FcγRI and is capable of ADCC (23). These cells had completely lost their proliferative capacities and attempts to increase their numbers using T-cell mitogens were not successful. Therefore, to harness their cytotoxic activity into a technology for use as an antitumor therapy, we initially cloned the native FcγRI into PBMCs from healthy donors. This receptor (named here A2G) is composed of the antibody-binding α-chain and the signaling γ-chain separated by a T2A skipping peptide and it was genetically engineered into peripheral blood T cells (Illustrated in Fig. 1A). Ectopic expression of fluorophore-tagged chains confirmed that they were expressed on the cell membrane (Supplementary Fig. S1A). Consistent with this, flow cytometry analysis of A2G-infected PBMCs showed that FcγRI receptor was highly expressed on T cells (Fig. 1B). To test the functionality of the receptor, we incubated transduced T cells with H2B-tdTomato labeled HT29 cells, which highly express HER2 antigen, with or without trastuzumab, and monitored tumor cell toxicity overtime using IncuCyte. Marked tumor cell lysis was observed only upon addition of trastuzumab, and only with A2G-transduced T cells, not with sham-transduced T cells (Fig. 1C and D). Nonetheless, as many tumor cells remained alive in culture, we tested whether the A2G construct could be improved. To this end, we fused an additional γ-chain to the C-terminus of the FcγRI α-chain (named AG2G; illustrated in Fig. 1A). In another construct (named AGO2GO), we further fused the signaling chains from OX40 to the C-terminus of the γ-chains (illustrated in Fig. 1A). The expression levels of both AG2G and AGO2GO in T cells were comparable with that of A2G (Fig. 1B; Supplementary Fig. S1B). Flow cytometry and qPCR analyses confirmed that both chains were equally expressed in human T cells (Supplementary Fig. S1C and S1D). Although the cytotoxic activity of T cells transduced with AG2G was significantly higher than that of A2G-transduced T cells, addition of OX40 signaling chains did not improve the cytotoxic activity of T cells (Fig. 1C and D; Supplementary Fig. S1E). Similarly, attempts to fuse other co-stimulatory signaling chains (i.e., CD28 and 4–1BB) showed no improved activity over T cells transduced with AG2G (Supplementary Fig. S1F). The addition of signaling motifs from costimulatory molecules induced non-specific activity upon incubation with irrelevant antibody or induced unattenuated levels of IFNγ (Supplementary Fig. S1G–S1J). Beyond IFNγ secretion, AG2G-transduced T cells selectively secreted IL2, TNFα, and GM-CSF, but not IL6, upon incubation with tumor-binding antibodies, but not with irrelevant antibodies (Fig. 1E). Taken together, while addition of the γ-chain to the FcγRI α-chain improved the ADCC activity of T cells transduced with the native FcγRI, addition of a co-stimulatory domain such as OX40, CD28, or 4–1BB was dispensable to this construct.
Figure 1.
Expression, cytotoxic activity, and cytokine secretion of T cells expressing altered version of FcγRI. A, Illustration of the modified FcγRI structure with fused signaling domains: A2G (left), FcγRI composed of α-chain and homodimer of FcRγ chain; AG2G (middle), FcγRI α-chain with fused domain of FcRγ chain and homodimer of FcRγ; AGO2GO (right), FcγRI α-chain with fused FcRγ and OX40 domains and homodimer of FcRγ with fused OX40 domain. B, Flow cytometry analysis of A2G, AG2G, and AGO2GO expression following retroviral transduction of healthy donor T cells. C, Mean numbers of HT29 target cells expressing HER2 calculated by incuCyte software through 48-hour incubation with the three FcγRI-based receptor expressing T cells, with 1:1 E:T ratios with or without 6 μg/mL trastuzumab (data shows one of two experiments repeats, n = 4). Each sample is normalized to its cell number at time zero. Statistical significance was calculated using two-way ANOVA with Sidak correction for multiple comparisons. D, Representative incuCyte images showing HER2-expressing HT29 cells, imaged and counted in live imaging system after 48 hours of incubation with different construct-expressing T cells with 1:1 E:T ratio. Scale bar is 100 μm. E, Human Luminex Discovery Assay measurement of cytokine levels in supernatants from 48 hours of culture of T cells 4:1 E:T ratio with HT29 cells expressing HER2 (n = 4). Graphs show mean ±SD. Statistical significance was calculated using two-way ANOVA with Tukey's correction for multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars represent standard error.
Characterization of activation and cytotoxicity of AG2G-expressing T cells
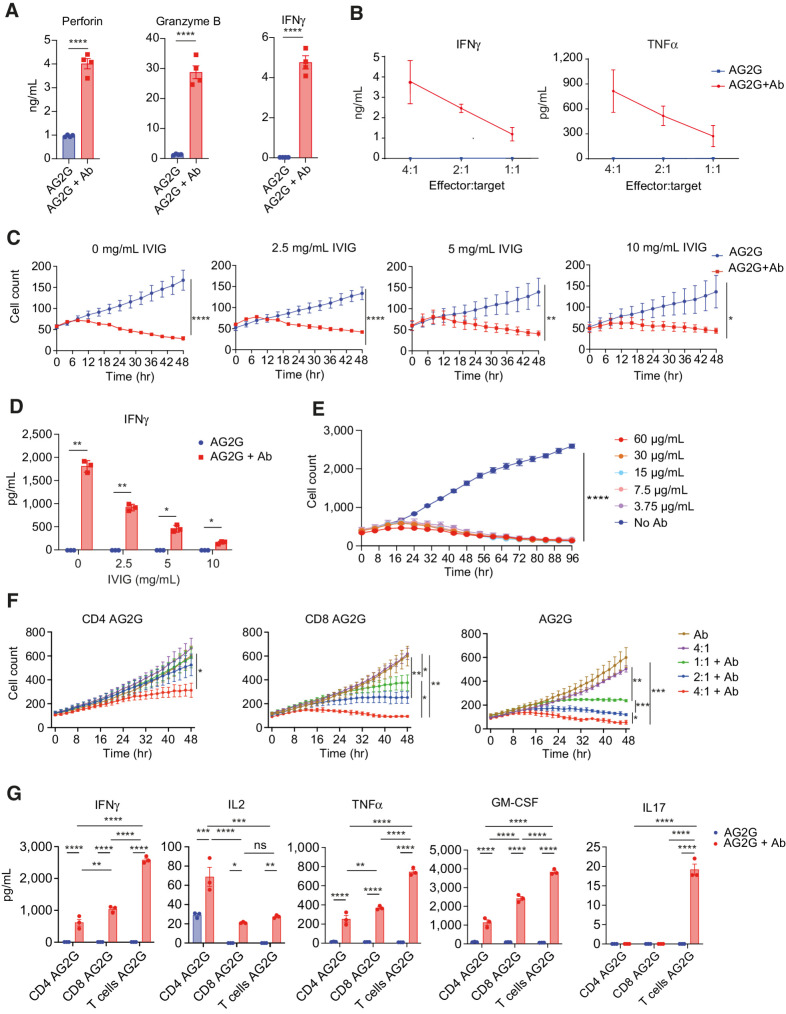
To further elucidate the killing mechanism of AG2G-expressing T cells, we tested if cytokine secretion was also associated with secretion of lytic granules. Hence, we measured the levels of perforin and granzyme B levels in the supernatants following coculture with the target cells, with or without trastuzumab. High levels of perforin, granzyme B and IFNγ were found only upon addition trastuzumab (Fig. 2A). Levels of secreted cytokines correlated to effector:target ratios (Fig. 2B). We also tested if killing rates were impaired in the presence of non-tumor binding antibodies. We found that the cytotoxic activity of AG2G was not inhibited by IVIG, although it did reduce IFNγ secreted from the activated cells (Fig. 2C and D). These results also demonstrated that no killing or cytokine secretion occurred upon incubation with circulating free antibodies. Titration of antibody concentration indicated an efficient killing activity that persisted for at least 4 days in range of 3.7 to 60 μg/mL trastuzumab (Fig. 2E).
Figure 2.
Characterization of AG2G-expressing T cells, activation, and cytotoxicity. A, ELISA measurement of cytokine levels of AG2G-expressing T cells after 48-hour incubation with HT29 cells expressing HER2. B, IFNγ and TNFα levels measured by ELISA in supernatants from 48-hour incubation of AG2G-expressing T cells and HT29 cells expressing HER2 in different E:T ratios. C, Mean numbers of HT29 cells expressing HER2, calculated by incuCyte software, following incubation with AG2G-expressing T cells in combination with 0, 2.5, 5, 10 mg/mL IVIG, with or without 30 μg/mL trastuzumab (n = 4). D, ELISA measurement of IFNγ levels of AG2G-expressing cells cocultured with HT29 cells expressing HER2 described in B. E, Mean numbers of HT29 cells expressing HER2, calculated by incuCyte software, following incubation with AG2G-expressing T cells in combination with different concentrations of trastuzumab over 96 hours (n = 2). F, Mean numbers of HT29 cells expressing HER2, calculated by incuCyte software, following incubation with different E:T ratios of AG2G-expressing T cells that were isolated for their CD4 (left) or CD8 (middle) population or with no isolation (right). G, ELISA measurement of cytokine levels from supernatants of 48-hour culture of 4:1 E:T ratios of isolated CD4, isolated CD8, or AG2G-expressing T cells with HT29 cells expressing HER2 and 30 μg/mL trastuzumab. Graphs show mean ±SD. Statistical significance was calculated using Student t test for a, two-way ANOVA with Sidak correction for multiple comparisons for B and C, two-way ANOVA with Tukey's correction for multiple comparisons for E and F. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars represent standard error.
Because AG2G-expressing T cells were composed of mixed CD8+ and CD4+ populations (Supplementary Fig. S2A), we next examined the contribution of CD4+ and CD8+ T cells to ADCC. AG2G-expressing T cells were separated into CD4+ and CD8+ cell populations (Supplementary Fig. S2B) and compared for their killing abilities. While expression of AG2G endowed CD4+ T cells with cytotoxic activity, the AG2G-expressing CD8 were more potent (Fig. 2F; Supplementary Fig. S2C). CD4+, CD8+, and mixed AG2G-T cells showed differential cytokine secretion upon addition of targeting antibody. While IL2 was predominantly secreted by CD4+ T cells, IFNγ and TNFα were secreted by both cell types in response to tumor-binding antibody, but they were higher in the mixed T cells (Fig. 2G). Th2 cytokines were relatively low and almost no IL6 was measured (Supplementary Fig. S2D). Overall, these data suggest that AG2G-transduced T cells respond only to tumor-bound, and not to free antibodies, and that both CD4+ and CD8+ T-cell subsets facilitate tumor ADCC.
To demonstrate the overreaching clinical therapeutic potential of AG2G-expressing T cells, we used them in combination with a number of different clinically approved IgG1 subtype antibodies. In addition to NCI-N87 targeting with trastuzumab (Supplementary Fig. S3A), other human tumor cell lines like JIMT-1 targeted with trastuzumab (Supplementary Fig. S3B), NCI-H1975 were targeted using anti-EGFR cetuximab (Supplementary Fig. S3C) or anti–PD-L1 avelumab (Supplementary Fig. S3D). A431 cells, which express EGFR, were also used to demonstrate targeting using necitumumab (Supplementary Fig. S3E), and HeLa cells were targeted with the anti-MUC16 sofituzumab (Supplementary Fig. S3F). Taken together these data establish AG2G-expressing T cells as a potential therapeutic platform enabling targeting of various TAAs.
AG2G-expressing T cells are not activated by low antigen level and avoid premature exhaustion
To further assess the specificity of ADCC activity of AG2G-expressing T cells, we compared the killing rates of HER2-expressing tumor cells in the presence of the anti-HER2 trastuzumab or irrelevant antibodies, rituximab and cetuximab. Killing of target cells was observed only upon incubation with AG2G-transduced T cells in combination with the correct targeting antibody, trastuzumab (Fig. 3A). Secretion of IFNγ by AG2G-expressing T cells was also observed only upon incubation with the antigen-specific, tumor-binding antibody trastuzumab, but not with the irrelevant antibodies (Fig. 3B).
Figure 3.
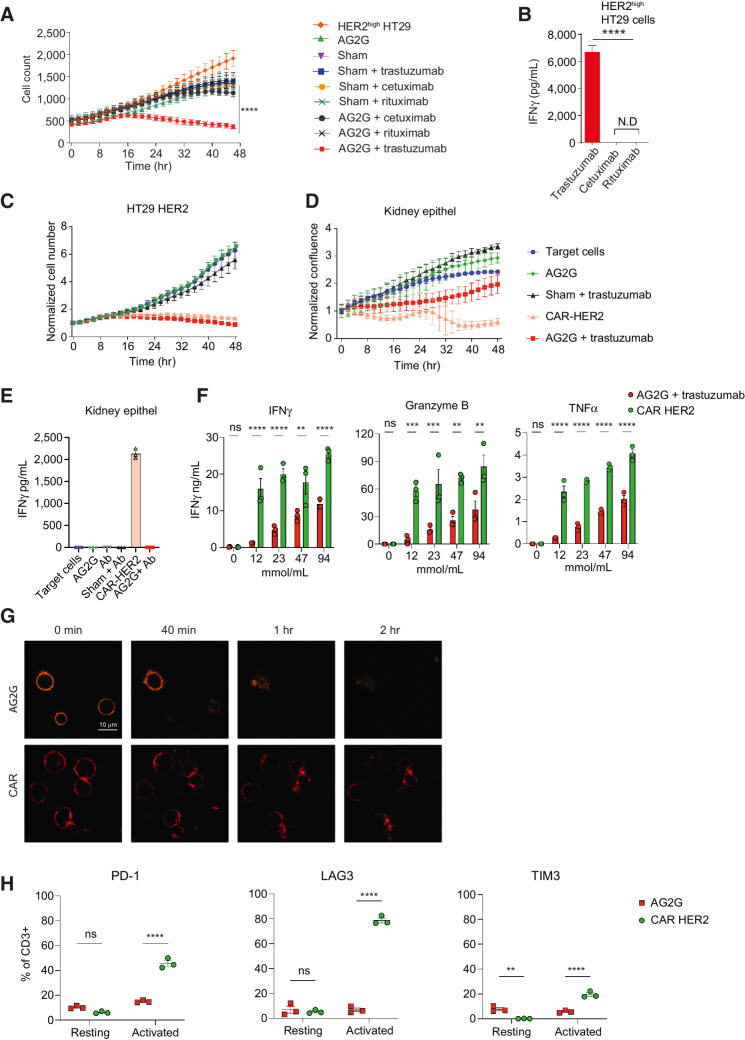
AG2G-expressing T cells differentiate between cells expressing high and low antigen levels, are more specific, and less exhausted, compared with classic CAR T cells. A, Mean numbers of HT29 cells expressing HER2, calculated by incuCyte software, following incubation with transduced T cells in combination with antibodies (60 μg/mL each, n = 4). B, ELISA measurement of IFNγ levels in 48-hour supernatants of AG2G-transduced T cells incubated with HER2-expressing HT29 target cells, and with tumor-binding trastuzumab, or irrelevant antibodies cetuximab and rituximab (60 μg/mL). C, Mean counts of HT29 cells expressing HER2, calculated by incuCyte software, following incubation with AG2G-transduced T cells and 30 μg/mL trastuzumab, compared with trastuzumab-derived CAR (4:1 E:T ratio, n = 4). Each sample is normalized to its cell number at time zero. D, Normalized confluence of kidney epithelial cells, calculated by incuCyte software, following incubation with AG2G-transduced T cells and 30 μg/mL trastuzumab, compared with trastuzumab-derived CAR (4:1 E:T ratio, n = 4). Each sample is normalized to its cell confluence at time zero. E, ELISA measurement of IFNγ levels in 48-hour supernatant obtained from transduced T cells and kidney epithel described in D (n = 4). F, ELISA measurement of IFNγ, granzyme B and TNFα levels in 24-hour supernatants of 105 AG2G-expressing T cells or CAR T cells incubated on immobilized trastuzumab or rHER2, in rising concentrations (mmol/mL) respectively. G, Confocal microscope imaging of internalization kinetics of PE-labeled IgG by AG2G-expressing cells (top), or PE-labeled HER2 by CAR T cells (bottom). H, PD-1, LAG-3, and TIM3 flow cytometry analysis of AG2G-expressing cells or CAR T cells after 48-hour incubation with or without 12 mmol/mL immobilized trastuzumab or HER2, respectively. Graphs show mean ± SD. Statistical significance was calculated using two-way ANOVA with Tukey's correction for multiple comparisons for A, C, D, and H. Two-way ANOVA with Sidak correction for multiple comparisons for E and F. One-way ANOVA with Dunnett correction for multiple comparisons for B. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars represent standard error.
Next, we sought to compare the potency and specificity of AG2G-expressing T cells with those of CAR T cells targeting the same antigen. To this end, we expressed a third-generation anti-HER2 CAR, with scFv derived from trastuzumab, in blood T cells, and tested their cytotoxic activity over time against HER2 overexpressing HT29 cells. The killing rates of AG2G and CAR transfected T cells were comparable across all time point (Fig. 3C). However, significant killing activity and IFNγ secretion were also observed when CAR T cells, but not AG2G-expressing T cells, were incubated with epithelial kidney cells that expressed physiologic baseline levels of HER2 (Fig. 3D and E).
The differential activity of AG2G-expressing T cells toward HER2 high and low expressing target cells, compared with CAR T cells, led us to test whether the density of ligand on targets affected the activity of these cells. Hence, we incubated transduced T cells with immobilized trastuzumab or recombinant HER2 and measured the level of IFNγ, TNFα, and granzyme B in overnight culture. In agreement with their killing properties, CAR T cells fully responded to different levels of immobilized HER2, whereas activation of AG2G-expressing T cells was dependent on levels of immobilized antibody (Fig. 3F). The correlation between density of immobilized antibody to activation of AG2G-expressing T cells, was repeated using other monoclonal IgG such as cetuximab or avelumab in addition to trastuzumab (Supplementary Fig. S4A). As tonic signaling was suggested to contribute to ligand-independent cytotoxicity in CAR T cells, we analyzed the expression patterns of AG2G and CAR receptors on the T-cell membrane. Confocal analysis demonstrated spontaneous clusters of HER2 CAR on the T-cell membrane, as opposed to the uniform patterns of AG2G (Supplementary Fig. S4B). In addition, incubation of transduced cells with corresponding fluorescent-labeled ligands showed that while IgG1 was internalized by the AG2G-expressing T cells within 40 minutes, soluble HER2 ligand was bound to HER2-CAR T cells for more than 2 hours (Fig. 3G). Whether these differences in sensitivity to the antigens affected the levels of T-cell exhaustion, was next tested. Analysis of PD-1, LAG-3, and TIM3 by flow cytometry of AG2G- and CAR T cells following 48-hour activation with immobilized trastuzumab or HER2 showed that the CAR T cells had higher levels of these exhaustion markers compared with AG2G-expressing T cells (Fig. 3H).
AG2G-expressing T cells combined with trastuzumab target HER2-expressing tumor cells, while sparing normal cells
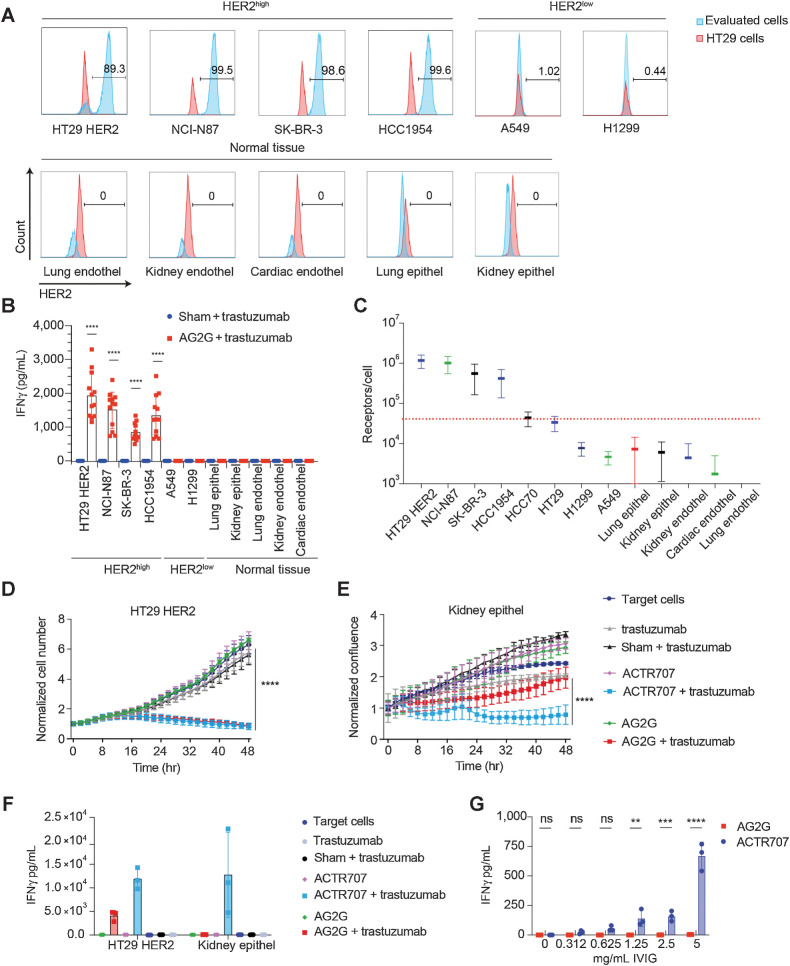
Given their dose-dependent activity, we next further assessed the ability of AG2G-expressing T cells to discriminate between tumor cells expressing high levels of antigens and a panel of normal cells with physiologic levels of HER2. We incubated AG2G-expressing T cells with tumor cells expressing high levels of HER2 (HT29-HER2, SK-BR3, Hcc1954, and NCI-N87), tumor cells with low expression of HER2 (A549, H1299), and normal primary lung and kidney epithelial, or lung, kidney, and cardiac endothelial cells (Fig. 4A; Supplementary Fig. S5A). T-cell activation, as manifested by IFNγ secretion, was only observed following incubation of AG2G-expressing T cells combined with trastuzumab, but not sham-transduced T cells, and only when incubated with target cells expressing high HER2 antigen density (Fig. 4B). No IFNγ secretion was observed in any of the primary cells expressing low levels of HER2 (Fig. 4B). Cytotoxic activity correlated with antigen density (Supplementary Fig. S5B). To determine the antigen density on cells that activate AG2G, we quantified the absolute antigen expression on different cell lines and correlated it to their killing by AG2G-expressing T cells. Our results indicated that approximately 40K receptors per cell were required to induce over 80% killing at an E:T ratio of 4:1 (Fig. 4C).
Figure 4.
Human T cells expressing AG2G exert specific tumor cytotoxicity while sparing normal cells. A, Flow cytometry expression analysis of HER2 on different tumor cell lines and primary human normal cells. B, ELISA measurement of IFNγ levels in 48-hour supernatants of AG2G-expressing T cells with different tumor cells or primary normal cells in 4:1 E:T ratio and 60 μg/mL trastuzumab (results shown are from 3 different donors combined, n = 12). C, Quantification of HER2 receptor numbers using DAKO QIFIKIT (Agilent) beads by flow cytometer. Individual populations of the calibration are gated, a linear regression is calculated from the MFI and the ABC values of the five calibration bead populations using MS Excel. Using the linear regression, the ABCs of the beads coated with αHER2 is calculated from their MFI values. SDs are also retrieved from gated populations and transformed using linear regression. D, Mean counts of HT29 cells expressing HER2, calculated by incuCyte software, following incubation with AG2G-transduced T cells or ACTR707 and 30 μg/mL trastuzumab (4:1 E:T ratio, n = 4). Each sample is normalized to its cell number at time zero. E, Normalized confluence of kidney epithelial (epithel) cells, calculated by incuCyte software, following incubation with AG2G-transduced T cells or ACTR707 and 30 μg/mL trastuzumab. Each sample is normalized to its cell confluence at time zero. Graphs are identical and described in Fig 3. C and D but with the comparison to ACTR707 instead of CAR T. F, ELISA measurement of IFNγ levels in 48-hour supernatant obtained from transduced T cells and kidney epithel described in D and E (n = 4). G, ELISA measurement of IFNγ levels in 24 hours supernatants of AG2G-expressing T cells or ACTR707 incubated with medium containing different concentration of IVIG (n = 3). Graphs show mean ±SD. Statistical significance was calculated using two-way ANOVA with Sidak correction for multiple comparisons. ****, P < 0.0001. Error bars represent standard error. Endothel, endothelial cells.
Genetic constructs such as ACTR707, which are composed of the extracellular domain of CD16 fused to CD28 and CD3z signaling chains have failed in the clinic due to toxicity. Hence, it was of interest to test their capacity to differentiate between high and physiologic levels of antigens. Similar to the results with anti-HER2 CAR T, ACTR707 T cells were highly active against target cells expressing high levels as well as physiologic levels of HER2 (Fig. 4D–F). Furthermore, ACTR707-transduced T cells were also activated by irrelevant antibodies, when added to HER2-expressing HT29 target cells (Supplementary Fig. S5D and S5E). Addition of free antibodies (IVIG) resulted in activation and secretion of IFNγ by ACTR707- but not AG2G-transduced T cells (Fig. 4G).
Taken jointly, our data strongly suggested that the basic scaffold of CAR T cells (also used by ACTR707) which fuses a number signaling chains, generates a construct that is too sensitive and cannot discriminate between normal and high antigen density.
Systemic administration of AG2G-expressing T cells in combination with trastuzumab eradicates HER2-expressing tumor cells in vivo
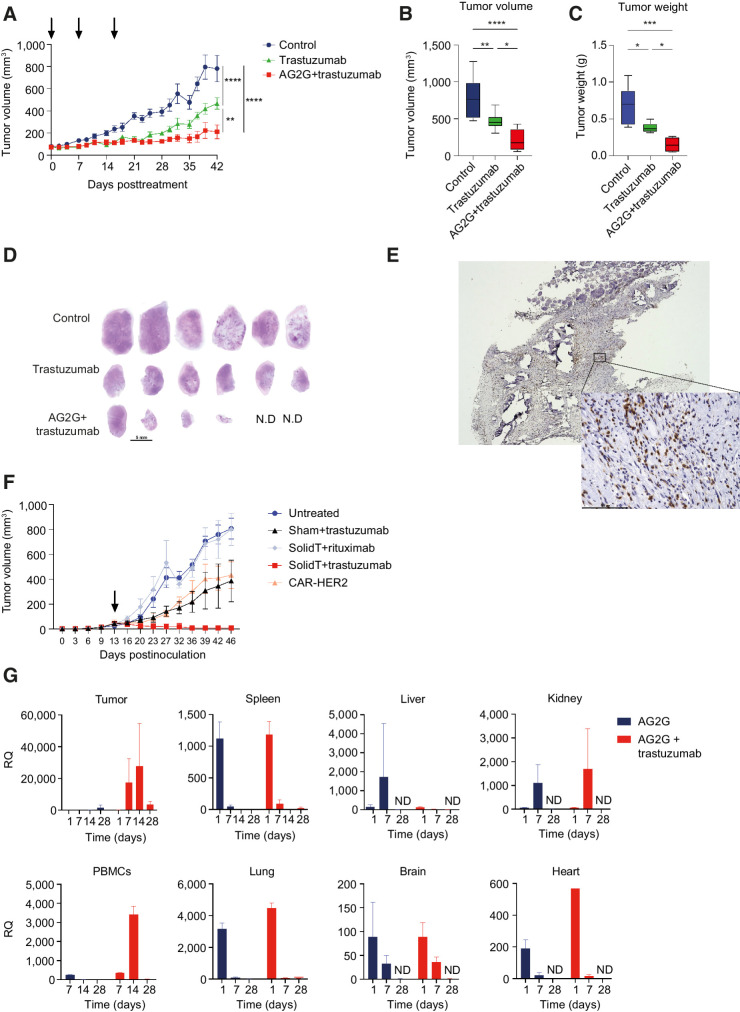
To assess the in vivo activity of AG2G-expressing T cells, we selected a gastric carcinoma xenograft model that endogenously expressed HER2 because this enabled the combination treatment with trastuzumab. 2.5×106 NCI-N87 cells were injected subcutaneously into NSG female mice. Once tumors reached 70 to 120 mm3, mice were randomized into three groups and treated once a week, for three weeks with intraperitoneal injection of trastuzumab and either intravenous injection of AG2G-expressing T cells (5×106) or saline, while control mice were treated with saline injection intraperitoneally and intravenously. Mice that received trastuzumab treatment alone or trastuzumab with AG2G-expressing T cells showed reduced tumor size. The addition of AG2G-expressing T cells, however, reduced tumor burden much more dramatically (Fig. 5A and B). Forty-two days following treatment initiation, mice were sacrificed and tumor weight was evaluated postmortem (Fig. 5C). The most significant reduction in tumor burden was observed in mice treated with the combination of trastuzumab and AG2G-expressing T cells. H&E staining of tumor sections at endpoint showed that the combination of trastuzumab and AG2G-expressing T cells resulted in total eradication of tumors in 2 mice of 6, while the others had smaller tumors compared with control mice and mice treated with trastuzumab alone (Fig. 5D). IHC staining with anti-human CD3, showed that of the six mice treated with AG2G-expressing T cells, four that had remaining tumor tissue after 42 days, had infiltration of human T cells into the tumors (Fig. 5E; Supplementary Fig. S6A). HER2 immunostaining of tumor samples at day 42 revealed that all tumors had a clinical IHC score of 3+ (Supplementary Fig. S6B). Histologic analysis of the tumor tissue revealed reduced necrosis score in the group treated with AG2G+trastuzumab (Supplementary Fig. S6C). Moreover, the AG2G+trastuzumab sample was the only one to show multiple mononuclear cells infiltrating intratumoral septa. No deviation from normal values was observed in any of the blood and chemistry parameters (Supplementary Fig. S6D) and body weight was not affected (Supplementary Fig. S6E). In addition, healthy organ histology revealed no abnormalities or toxicities in any of the mice following treatment with AG2G-expressing T cells (Supplementary Table S3).
Figure 5.
Systemic administration of AG2G-expressing T cells in combination with trastuzumab eradicates HER2-expressing tumor cells in vivo. A, NCI-N87 tumor volume measured by caliper over 42 days following treatment initiation. NSG mice were injected with 2.5×106 NCI-N87 tumor cells, after tumor reached 70 mm3 mice were randomized and treated once a week for 3 times (treatments are marked with arrows) with either intraperitoneal injection of saline or 250-μg trastuzumab and intravenous injection of saline or 5×106 AG2G-expressing T cells (n = 6). B, Endpoint analysis of tumor volume measured by caliper, 42 days following treatment initiation. C, Endpoint analysis of tumor weight measured using scales, 42 days following treatment initiation. D, Histology images taken by x4 objective light microscopy, showing whole sections of tumors stained with H&E (n = 6; N.D., not detected - 2 mice from AG2G+trastuzumab group). Scale bar is 5 mm. E, Representative image taken by x4 and x40 objectives using light microscopy, of one tumor from AG2G+trastuzumab group stained with anti-human CD3 antibody by IHC. F, Tumor volume measured by caliper in NSG mice treated with a single injection of T cells (107) 13 days after tumor inoculation (2.5×106 NCI-N87). Antibodies were injected once a week for a total of 3 times (250 μg/mouse), n = 5. G, NSG mice were injected with 1.7×106 NCI-N87 tumor cells, after tumor reached 100 to 200 mm3 mice were randomized and treated once with intravenous injection of 5×106 AG2G-expressing T cells and with intraperitoneal injection of saline or 250-μg trastuzumab. On days 1, 7, 14, 28 following treatment, 3 mice from each group were sacrificed and RNA was extracted from blood and tumors for real-time PCR analysis of AG2G-expressing cells (n = 12). Graphs show mean ± SD. Graphs A–C show one of two independent experiments performed. Statistical significance was calculated using two-way ANOVA with Tukey's correction for multiple comparisons for A and B, one-way ANOVA with Holm-Sidak correction for multiple comparisons for C. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars represent standard error.
We next compared the efficacy of AG2G-transduced T cells to that of HER2-CAR T cells against HER2. Mice bearing established NCI-N87 tumors were treated with a single dose of 1×107 transduced T cells. We found that while administration of AG2G-transdued T cells with irrelevant antibody (i.e., rituximab) was almost inert, administration of AG2G plus trastuzumab completely cleared the tumors in all treated mice. Mice treated with sham-transfected or with CAR T cells (Supplementary Fig. S6F), showed comparable tumor growth inhibition, but none of the tumors were cleared (Fig. 5F; Supplementary Fig. S6G).
Next, we injected a single dose of AG2G-expressing T cells, with or without trastuzumab, to follow cell infiltration and persistence in tumors. qPCR of the AG2G receptor showed that the cells were detected in blood of naïve mice after their injection, and their numbers were lowered, until not detected by day 14 after the injection (Supplementary Fig. S6H). In tumor-bearing mice AG2G-expressing cell detection was much higher when antibody was injected (Fig. 5G), and while their numbers were reduced in blood over time, they accumulated in the tumors, where they persisted for at least 28 days following injection (Fig. 5G; Supplementary Fig. S6I). These data highly support the potential of AG2G-expressing T cells as an efficacious and safe T-cell therapy targeting solid tumors.
Systemic administration of tumor-binding antibodies and AG2G-expressing T cells eradicates melanoma in vivo
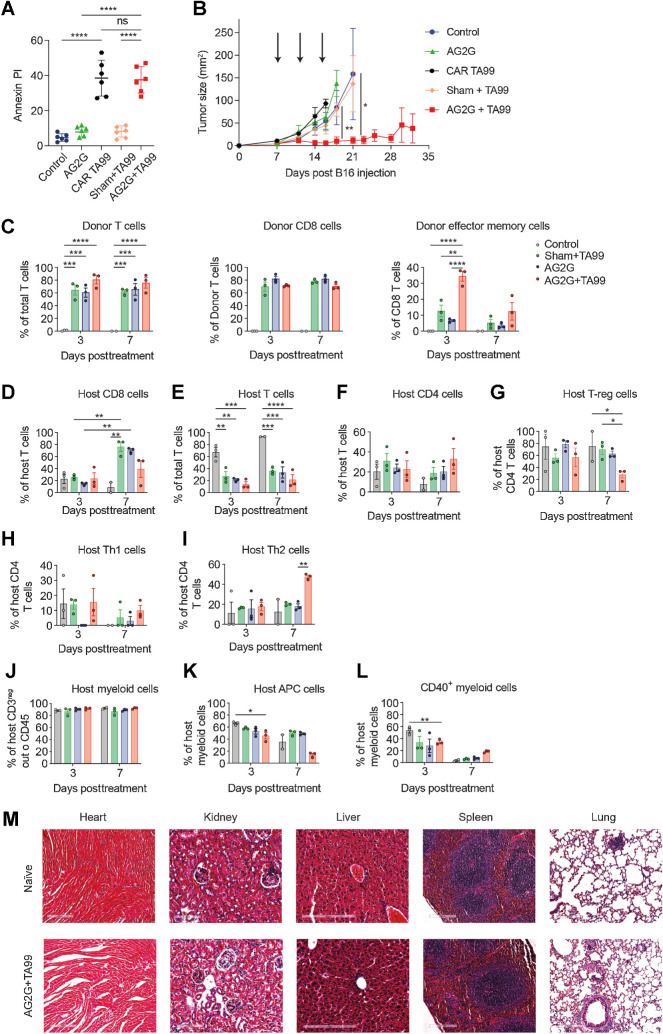
Next, we evaluated the efficacy and function of AG2G-expressing T cells in an immunocompetent C57Bl/6 mice B16F10 melanoma model in which the tumor can be targeted with the anti–TRP-1/TA99 antibody, which can be used with mouse AG2G-expressing T cells. We compared the capacity of AG2G-transduced T cells to eradicate palpable tumors to that of sham transduced T cells as well as T cells expressing a CAR derived from TA99 (24). Initially, we tested their capacity to kill B16F10 tumors in vitro. We found that at an E:T ratio of 4:1 both CAR and AG2G-transduced T cells induced approximately 40% tumor cell lysis (Fig. 6A). Next, C57BL/6 mice were injected subcutaneously with B16F10 melanoma cells, and tumors were allowed to grow for 7 days until they reached palpable size. Mice were then injected intravenously 3 times, 2 days apart, with sham-transduced T cells, with TA99-derived CAR-transduced T cells, or with mouse T cells expressing the AG2G construct, in combination with the TA99 antibody. Treatment with sham-transduced T cells combined with TA99 or with TA99-derived CAR T cells was largely ineffective, and tumor growth was comparable with that of untreated mice (Fig. 6B). In contrast, treatment with AG2G-transduced T cells combined with TA99 antibodies, but not AG2G-transduced T cells alone, induced remarkable tumor regression in all mice and resulted in complete tumor clearance in 50% of the treated mice (Fig. 6B). The therapeutic effect of AG2G-transduced T cells might be induced directly, through grafted T-cell infiltration, or through induction of host T-cell immunity.
Figure 6.
Systemic administration of tumor-binding antibodies and T cells expressing AG2G eradicate established melanoma. A, Annexin-V/PI positive target cells after 16 hours of B16F10 coculture and mouse sham untransduced T cells, CAR T derived from TA99, or AG2G transduced cells with or without 60 μg/mL TA99 antibody (4:1 E:T ratio). n = 6. B, Tumor size following intravenous injection of 1×106 splenic T cells transduced with the indicated vector, with or without subcutaneous injection of 250 μg/mouse of TA99 antibodies (treatments are indicated by arrows, n = 4). C, Bar graph representation of CyTOF analysis of CD45.1 donor T cells (CD3), CD8 T cells or effector memory cells (PD-1-/CD69+/CD44+/CD62L-/CD8+ cells). n = 3. D–L, Bar graph representation of CyTOF analysis and CD45.2 host infiltrate in B16F10 tumors 3 and 7 days posttreatment with sham untransduced or AG2G-transduced T cells with or without TA99 targeting antibody or control untreated mice. T cells were defined as CD45+/CD3+/TCRβ, myeloid cells were defined as CD45+/CD11b+/CD3neg/CD19neg/NK1.1neg, APC were defined as myeloid cells but with the addition of MHCII. n = 3. M, Images taken by x20 objective using slide scanner of H&E staining of histologic sections of naïve mice and of tumor-bearing mice treated with AG2G-transduced T cells and TA99 antibodies described in B. Graphs show mean ±SD. Statistical significance was calculated using two-way ANOVA with Tukey's correction for multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars represent standard error.
To further study whether the therapeutic effect of AG2G stems from infiltration of the transduced T cells or host-derived cells into the tumor tissue, we used CD45.2 host C57BL/6 mice bearing B16F10 melanoma tumors and treated them with AG2G-expressing or sham-transfected T cells that originated from CD45.1 donor mice, with or without the addition of tumor-binding antibody (TA99). Tumors were dissected on different days after treatment and subjected to CyTOF analysis. Comparable levels of donor T cells were detected in tumors of all groups. Nonetheless, only mice treated with AG2G-transduced T cells plus TA99 antibody had infiltrating T cells expressing activation and memory markers CD69, CD44, CD62 L, and T-bet in tumors (Fig. 6C; Supplementary Fig. S7A and S7B). In addition, increased infiltration of host CD45.2 T cells, mainly CD4+ T cells, was observed when AG2G-expressing T cells plus TA99 antibody were administered. Exhaustion markers such as PD-1 were not highly expressed in both CD4+ and CD8+ infiltrating cells, yet were more abundant on host CD4+ cells (Fig. 6D–I). Furthermore, the tumor infiltrate consisted of a large myeloid population of which a specific population of APCs with high CD40 expression was found to be increased in tumors from mice administered AG2G-expressing T cells plus TA99 antibody (Fig. 6J–L; Supplementary Fig. S7C–S7I). Finally, postmortem analysis did not indicate any obvious immune-cell infiltration into off-target tissues such as the heart, kidneys, liver, spleen, and lungs (Fig. 6M; Supplementary Fig. S7J).
Retroviral transduction of γδ-T cells with AG2G endows them with antitumor ADCC capacity
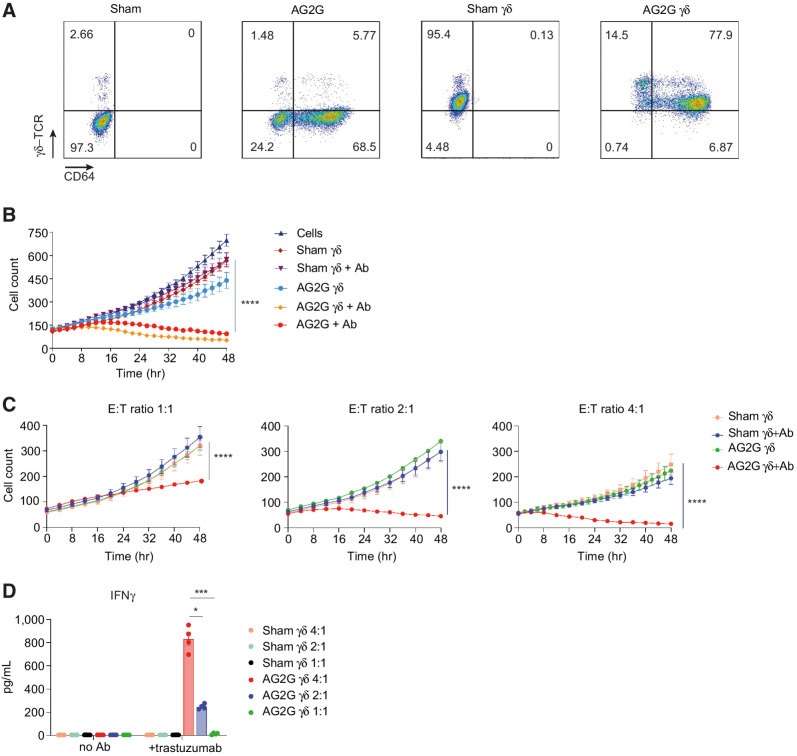
We next asked whether AG2G would be active in γδ-T cells. To this end, PBMCs were stimulated with zoledronic acid prior to transduction with AG2G encoding retrovirus (25). Flow cytometry analysis shows that over 90% of produced cells were γδ-T cells, of them about 80% were transduced with the AG2G construct (Fig. 7A). Using HT29-HER2 target cells expressing H2b-tdTomato, we showed that AG2G-transduced γδ-T cells killed the tumor cells upon addition of trastuzumab in a similar manner to αβ T cells (Fig. 7B). Similar patterns of tumor-cell killing and IFNγ expression were also observed using NCI-N87 cells line, which naturally express high density of HER2 (Supplementary Fig. S8A and S8B). Importantly, no major contamination of α/β T cells was observed in any of the samples even after 48 hours in culture (Supplementary Fig. S8C and S8D). Similar to our results with α/β T cells, the cytotoxic response of transduced γδ-T cells and their secretion of IFNγ correlated with E:T ratios, and a potent cytotoxic response was obtained with E:T ratios as low as 4:1, 2:1 and 1:1 (Fig. 7C and D). These results open the possibility for future development of allogeneic AG2G-expressing T cells as a platform cell therapy technology.
Figure 7.
Retroviral transduction of γδ-T cells with AG2G endows them with antitumor ADCC. A, Flow cytometry analysis of sham or AG2G-transduced αβ-T cells (two left panels, activated with IL2 and anti-CD3); sham or AG2G-transduced γδ-T cells (two right panels, activated with IL2 and zoledronic acid). B, Live cell numbers of HT29-HER2 expressing cells, calculated by incuCyte software, cocultured with AG2G-expressing γδ-T cells or AG2G-expressing αβ-T cells, with or without trastuzumab (n = 3). C, Live cell numbers of HT29-HER2 expressing cells, calculated by incuCyte software, cocultured with sham untransduced or AG2G-expressing γδ-T cells with or without trastuzumab, in E:T ratios of 1:1, 2:1 or 4:1 (n = 4). D, ELISA measurement of IFNγ levels in supernatants of 48-hour sham or AG2G-expressing γδ-T cells cocultured with HT29-HER2 expressing cells, with or without trastuzumab (E:T ratio 4:1, n = 3). Graphs show mean ±SD. Statistical significance was calculated using two-way ANOVA with Tukey's correction for multiple comparisons for B and C. Two-way ANOVA with Sidak correction for multiple comparisons for D. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Error bars represent standard error.
Discussion
Because the clinical demonstration that anti-CD19 CAR T cells can effectively induce durable remission of refractory chronic lymphoblastic leukemia (26, 27), tremendous research efforts have been exerted to replicate this success in other malignancies. Subsequently, similar response rates and long-term remission were also observed in refractory lymphoblastic leukemia (28), non-Hodgkin lymphoma, and diffuse large B-cell lymphoma (29). More recently, CAR T-cell therapy targeting BCMA, has shown promising results in multiple myeloma (30) and possibly in acute myeloid lymphoma (31). Disappointingly, however, attempts to date to employ CAR T-cell therapy to solid cancers have been largely unsuccessful (32, 33). Several theories have been proposed to explain the differential response rate between CAR T-cell therapy in solid tumors and hematologic malignancies. The most accepted hypothesis is that the suppressive tumor microenvironment and the harsh metabolic conditions that exist in solid and dense tumors prevent CAR T cells from infiltrating into the tumors and result in rapid exhaustion (34–36).
Multiple strategies have been developed to overcome these limitations, including expression of chemokine receptors (e.g., CCR2) to increase CAR T-cell migration (37), tumor-induced cytokine secretion to support CAR activation (38), and metabolic modifications to increase cell survival (39). The fact that these strategies are insufficient to induce regression in solid tumors, is somewhat surprising, as administration of tumor-infiltrating lymphocytes has been shown to induce tumor remission in similar clinical settings and preclinical models (5, 40). Alternatively, other studies have suggested that the spatial architecture of ScFv and the high-affinity nature of their interactions with ligands is what limits the clinical efficacy of CAR T cells in solid tumors. Interestingly, replacing the ScFv with a TCR and fusing it to co-stimulatory receptors has been shown to induce faster and more prominent T-cell infiltration, followed by more robust antitumor immunity (38, 41).
We show that AG2G-expressing T cells massively infiltrated into tumors in both NSG and immunocompetent mouse models and adopted an activated phenotype leading to tumor eradication. This is possible because unlike CAR T cells they do not upregulate checkpoint blockade molecules in response to low amounts of antigens expressed on normal cells, thus making them less susceptible to inhibition at the tumor microenvironment. In most cases however, it is the severe adverse toxicity, rather than insufficient signaling, that limits CAR T-cell activity (32, 42). One extreme example is the case of anti-HER2 CAR T-cell therapy where a treated patient died as a result of this therapy (43). Our data support the finding that anti-HER2 CAR T cells kill also healthy cells that express physiologic HER2 levels. Confocal analyses indicated that the anti-HER2 CAR molecules and CD3ζ molecules formed aggregates in transfected T cells, which are likely to lead to induction of tonic signaling, resulting in premature T-cell exhaustion. In a seminal paper, Salter and colleagues have shown that in CD19-CAR T-cell therapy, LCK is constitutively bound to CD28 and to a lesser extent also to 4–1BB. As a result, most of the TCR phosphorylation sites are activated in both unstimulated and stimulated CD19-CAR T cells (44). Consistent with this, Long and colleagues have shown that a number of CARs (including the 4D5, used here to target HER2) tend to cluster due to framework regions in ScFv, leading to tonic signaling, CD3ζ phosphorylation and early T-cell exhaustion (45).
AG2G is constructed such that the tumor-binding chain is separated from the signaling chains, thus allowing induction of high-affinity tumor-recognition receptors while limiting the response only to high antigen density. This fundamental change enables AG2G-transduced T cells to discriminate between normal and cancerous cells expressing the same antigen. Coupling the signaling chains to high recognition receptor as in most CAR T design, generates a receptor that is too sensitive to discriminate between normal and cancerous cells as several hundred receptors expressed on the target cells are sufficient induce T-cell activation (46). While this feature is extremely useful to eradicate lymphomas, in which normal B cells are depleted, it manifests a huge problem when treating solid epithelial cells. In the case of HER2 for example, normal tissues cells express about 10,000 receptors per cell. As a result, attempts to treat patients with HER2+ cancers were highly toxic (43). Clémenceau and colleagues were the first to employ a therapeutic strategy in cancer by expressing CD16158V fused to the FcεRIγ signaling chain in CD8+ T cells (47). While transduced T cells showed in vitro killing of antibody-coated B cells, the E:T ratios were very high (30:1) and no in vivo models were further treated. The premise behind that construct is the generation of heterodimer with CD3ζ. Because it occurs specifically in NK cells, subsequent translation of this strategy was executed using NK92 cells. An alternative strategy has been proposed by Kudo and Caratelli who built upon conventional CAR design by fuseing CD16158V or CD32A131R respectively, to 4–1BB (or CD28) followed by the CD3ζ signaling chains (48, 49). While these constructs showed promising results in preclinical models, they have failed in the clinic, inducing the death of cancer patients in two cohorts. Here, we demonstrate that such constructs are activated by free circulating antibodies, resulting in killing of normal cells expressing physiologic levels of antigens. While AG2G-transduced T cells avoided that, it may still risk individuals with active autoimmunity, or chronic inflammatory diseases associated with high titer of self-recognizing antibodies. Therapeutic strategies that are based on separating the tumor-binding entity and the T-cell signaling, avoid nonspecific activation by circulating antibodies. These strategies include tumor-binding molecule (such as, Aza, Folate, or tumor-binding antibody) fused to CAR T engager or adaptor molecules (i.e., FITC, or E5B9 peptide) and are administrated along with CAR T cells against the adaptor/engager molecules [reviewed in (50, 51)]. While these technologies suggest simpler implementations of logic gates, our results suggest that the hypersensitivity structure of the CAR design in the T cells may limit their potency due to binding to free engager/adaptor. In addition, as opposed to antibodies, chimeric molecules are likely to have a short half-life in the circulation and even stimulate production of antibodies against them.
In conclusion, our work demonstrates the capacity of a modified FcγRI transduced into T cells and combined with tumor-binding antibody, to eliminate solid tumors. We believe that our results establish the basis for a novel signal initiation scaffold in which the tumor-binding entity is not coupled with the T-cell signaling, which enables T cells to decipher between normal and cancerous cells based on their expression of antigen density. Moreover, this construct provides technological means to treat different tumor types using a single foundational T-cell product and without changing the manufacturing program. Beyond this technology's ability to treat a wide range of solid cancers, this design can actively target refractory tumors that reduce their antigen expression by replacing the injected antibody or combining it with additional binding antibodies. Finally, it is a platform to repurpose antibodies that efficiently bind tumor cells but fail to show clinical efficacy.
Supplementary Material
Supplementary tables and figures
Acknowledgments
This work was funded by the Emerson Collective Foundation, the ICRF, the Dotan Foundation, and the Israel Science Foundation (ISF; grant number 2262/18).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
D. Rasoulouniriana reports personal fees from Gilboa Therapeutics during the conduct of the study; in addition, D. Rasoulouniriana has a patent for WO/2020/188570 pending and licensed, a patent for WO/2021/260696 pending and licensed, a patent for 63/435,352 pending and licensed, and a patent for 63/439,608 pending and licensed. S. Amar reports grants from Gilboa Therapeutics during the conduct of the study. H. Shpilt reports personal fees from Gilboa Therapeutics during the conduct of the study; in addition, H. Shpilt has a patent for 63/439,608 pending and licensed to Gilboa Therapeutics and a patent for 63/435,352 pending and licensed to Gilboa Therapeutics. S. Dotan reports personal fees from Gilboa Therapeutics during the conduct of the study; personal fees from Gilboa Therapeutics outside the submitted work; in addition, S. Dotan has a patent for 63/435,352 pending to Gilboa Therapeutics and a patent for 63/439,608 pending to Gilboa Therapeutics. N. Pilpel reports personal fees from Gilboa Therapeutics during the conduct of the study; personal fees from Gilboa Therapeutics outside the submitted work; in addition, N. Pilpel has a patent for 63/435,352 pending and licensed to Gilboa Therapeutics and a patent for 63/439,608 pending and licensed to Gilboa Therapeutics. P. Rider reports personal fees and other support from Gilboa Therapeutics during the conduct of the study; in addition, P. Rider has a patent for WO/2020/188570 pending and licensed, a patent for WO/2021/260696 pending and licensed, a patent for 63/435,352 pending and licensed, and a patent for 63/439,608 pending and licensed. Y. Carmi reports grants and other support from Gilboa Therapeutics during the conduct of the study; other support from Bolt Biotherapeutics outside the submitted work; in addition, Y. Carmi has a patent for wo/2020/188570 pending and licensed to Gilboa Therapeutics, a patent for wo/2021/260696 pending and licensed to Gilboa Therapeutics, a patent for 63/435,352 pending and licensed to Gilboa Therapeutics, and a patent for 63/439,608 pending and licensed to Gilboa Therapeutics. No disclosures were reported by the other authors.
Authors' Contributions
D. Rasoulouniriana: Investigation, methodology. N. Santana-Magal: Investigation. A. Gutwillig: Software, visualization. L. Farhat-Younis: Investigation, methodology. L. Tal: Investigation. S. Amar: Investigation. M. Milyavsky: Resources. S.S.N.A. Muddineni: Resources. N. Solomon: Investigation. H. Shpilt: Investigation, methodology. S. Dotan: Investigation, writing–original draft, writing–review and editing. N. Pilpel: Supervision, writing–original draft, writing–review and editing. C. Waskow: Resources. M. Feinmesser: Resources, methodology. P. Rider: Investigation, writing–review and editing. Y. Carmi: Supervision, investigation, writing–original draft, writing–review and editing.
References
- 1. Apperley JF, Jones L, Hale G, Waldmann H, Hows J, Rombos Y, et al. Bone marrow transplantation for patients with chronic myeloid leukemia: T-cell depletion with Campath-1 reduces the incidence of graft-versus-host disease but may increase the risk of leukemic relapse. Bone Marrow Transplant 1986;1:53–66. [PubMed] [Google Scholar]
- 2. Marmont AM, Horowitz MM, Gale RP, Sobocinski K, Ash RC, van Bekkum DW, et al. T-cell depletion of HLA-identical transplants in leukemia. Blood 1991;78:2120–30. [PubMed] [Google Scholar]
- 3. Pollard CM, Powles RL, Millar JL, Shepherd V, Milan S, Lakhani A, et al. Leukemic relapse following Campath 1 treated bone marrow transplantation for leukemia. Lancet 1986;2:1343–4. [DOI] [PubMed] [Google Scholar]
- 4. Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T-cell response. Nat Rev Immunol 2012;12:269–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res 2011;17:4550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu R, Forget MA, Chacon J, Bernatchez C, Haymaker C, Chen JQ, et al. Adoptive T-cell therapy using autologous tumor-infiltrating lymphocytes for metastatic melanoma: current status and future outlook. Cancer J 2012;18:160–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anguille S, Van Tendeloo VF, Berneman ZN. Leukemia-associated antigens and their relevance to the immunotherapy of acute myeloid leukemia. Leukemia 2012;26:2186–96. [DOI] [PubMed] [Google Scholar]
- 8. Inoue K, Ogawa H, Sonoda Y, Kimura T, Sakabe H, Oka Y, et al. Aberrant overexpression of the Wilms tumor gene (WT1) in human leukemia. Blood 1997;89:1405–12. [PubMed] [Google Scholar]
- 9. Krackhardt AM, Witzens M, Harig S, Hodi FS, Zauls AJ, Chessia M, et al. Identification of tumor-associated antigens in chronic lymphocytic leukemia by SEREX. Blood 2002;100:2123–31. [DOI] [PubMed] [Google Scholar]
- 10. Garber HR, Mirza A, Mittendorf EA, Alatrash G. Adoptive T-cell therapy for leukemia. Mol Cell Ther 2014;2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wagner JE, Thompson JS, Carter SL, Kernan NA; Unrelated Donor Marrow Transplantation T. Effect of graft-versus-host disease prophylaxis on 3-year disease-free survival in recipients of unrelated donor bone marrow (T-cell depletion trial): a multicenter, randomized Phase II-III trial. Lancet 2005;366:733–41. [DOI] [PubMed] [Google Scholar]
- 12. Palucka AK, Coussens LM. The basis of oncoimmunology. Cell 2016;164:1233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sadelain M, Brentjens R, Riviere I. The basic principles of chimeric antigen receptor design. Cancer Discov 2013;3:388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barrett DM, Grupp SA, June CH. Chimeric antigen receptor- and TCR-modified T cells enter main street and wall street. J Immunol 2015;195:755–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garfall AL, Maus MV, Hwang WT, Lacey SF, Mahnke YD, Melenhorst JJ, et al. Chimeric antigen receptor T cells against CD19 for multiple myeloma. N Engl J Med 2015;373:1040–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 2014;371:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gross G, Eshhar Z. Therapeutic potential of T cell chimeric antigen receptors (CARs) in cancer treatment: counteracting off-tumor toxicities for safe CAR T-cell therapy. Annu Rev Pharmacol Toxicol 2016;56:59–83. [DOI] [PubMed] [Google Scholar]
- 18. Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. The future of cancer treatment: immunomodulation, CARs, and combination immunotherapy. Nat Rev Clin Oncol 2016;13:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gardner R, Wu D, Cherian S, Fang M, Hanafi LA, Finney O, et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR T-cell therapy. Blood 2016;127:2406–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maude S, Barrett DM. Current status of chimeric antigen receptor therapy for hematologic malignancies. Br J Haematol 2016;172:11–22. [DOI] [PubMed] [Google Scholar]
- 21. Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov 2015;5:1282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ajina A, Maher J. Strategies to address chimeric antigen receptor tonic signaling. Mol Cancer Ther 2018;17:1795–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rasoulouniriana D, Santana-Magal N, Gutwillig A, Farhat-Younis L, Wine Y, Saperia C, et al. A distinct subset of FcγRI-expressing Th1 cells exert antibody-mediated cytotoxic activity. J Clin Invest 2019;129:4151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma L, Dichwalkar T, Chang JYH, Cossette B, Garafola D, Zhang AQ, et al. Enhanced CAR T-cell activity against solid tumors by vaccine boosting through the chimeric receptor. Science 2019;365:162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rozenbaum M, Meir A, Aharony Y, Itzhaki O, Schachter J, Bank I, et al. Gamma-delta CAR T cells show CAR-directed and independent activity against leukemia. Front Immunol 2020;11:1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med 2013;5:177ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med 2011;3:95ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric antigen receptor—modified T cells for acute lymphoid leukemia. N Engl J Med 2013;368:1509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, et al. CD22-targeted CAR T cells induce remission in B-ALL that is naïve or resistant to CD19-targeted CAR immunotherapy. Nat Med 2018;24:20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ali SA, Shi V, Maric I, Wang M, Stroncek DF, Rose JJ, et al. T cells expressing an anti–B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood 2016;128:1688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cummins KD, Gill S. Will CAR T-cell therapy have a role in AML? Promises and pitfalls. Semin Hematol 2019;56:155–63. [DOI] [PubMed] [Google Scholar]
- 32. Hou AJ, Chen LC, Chen YY. Navigating CAR-T cells through the solid-tumor microenvironment. Nat Rev Drug Discov 2021;20:531–50. [DOI] [PubMed] [Google Scholar]
- 33. Newick K, O'Brien S, Moon E, Albelda SM. CAR T-cell therapy for solid tumors. Annu Rev Med 2017;68:139–52. [DOI] [PubMed] [Google Scholar]
- 34. Anderson KG, Stromnes IM, Greenberg PD. Obstacles posed by the tumor microenvironment to T-cell activity: a case for synergistic therapies. Cancer Cell 2017;31:311–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joyce JA, Fearon DT. T-cell exclusion, immune privilege, and the tumor microenvironment. Science 2015;348:74–80. [DOI] [PubMed] [Google Scholar]
- 36. Lim AR, Rathmell WK, Rathmell JC. The tumor microenvironment as a metabolic barrier to effector T cells and immunotherapy. Elife 2020;9:e55185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moon EK, Carpenito C, Sun J, Wang LC, Kapoor V, Predina J, et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin Cancer Res 2011;17:4719–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baeuerle PA, Ding J, Patel E, Thorausch N, Horton H, Gierut J, et al. Synthetic TRuC receptors engaging the complete T-cell receptor for potent antitumor response. Nat Commun 2019;10:2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu X, Gnanaprakasam JNR, Sherman J, Wang R. A metabolism toolbox for CAR T therapy. Front Oncol 2019;9:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bernhard H, Neudorfer J, Gebhard K, Conrad H, Hermann C, Nahrig J, et al. Adoptive transfer of autologous, HER2-specific, cytotoxic T lymphocytes for the treatment of HER2-overexpressing breast cancer. Cancer Immunol Immunother 2008;57:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumor rejection. Nature 2017;543:113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rafiq S, Hackett CS, Brentjens RJ. Engineering strategies to overcome the current roadblocks in CAR T-cell therapy. Nat Rev Clin Oncol 2020;17:147–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther 2010;18:843–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Salter AI, Ivey RG, Kennedy JJ, Voillet V, Rajan A, Alderman EJ, et al. Phosphoproteomic analysis of chimeric antigen receptor signaling reveals kinetic and quantitative differences that affect cell function. Sci Signal 2018;11:eaat6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, et al. 4-1BB costimulation ameliorates T-cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med 2015;21:581–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heitzeneder S, Bosse KR, Zhu Z, Zhelev D, Majzner RG, Radosevich MT, et al. GPC2-CAR T cells tuned for low antigen density mediate potent activity against neuroblastoma without toxicity. Cancer Cell 2022;40:53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clemenceau B, Congy-Jolivet N, Gallot G, Vivien R, Gaschet J, Thibault G, et al. Antibody-dependent cellular cytotoxicity (ADCC) is mediated by genetically modified antigen-specific human T lymphocytes. Blood 2006;107:4669–77. [DOI] [PubMed] [Google Scholar]
- 48. Kudo K, Imai C, Lorenzini P, Kamiya T, Kono K, Davidoff AM, et al. T lymphocytes expressing a CD16 signaling receptor exert antibody-dependent cancer cell killing. Cancer Res 2014;74:93–103. [DOI] [PubMed] [Google Scholar]
- 49. Caratelli S, Sconocchia T, Arriga R, Coppola A, Lanzilli G, Lauro D, et al. FCγ chimeric receptor-engineered T cells: methodology, advantages, limitations, and clinical relevance. Front Immunol 2017;8:457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arndt C, Fasslrinner F, Loureiro LR, Koristka S, Feldmann A, Bachmann M. Adaptor CAR platforms-next generation of T cell–based cancer immunotherapy. Cancers (Basel) 2020;12:1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin H, Cheng J, Mu W, Zhou J, Zhu L. Advances in universal CAR T-cell therapy. Front Immunol 2021;12:744823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables and figures
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data files or upon request from the corresponding author.