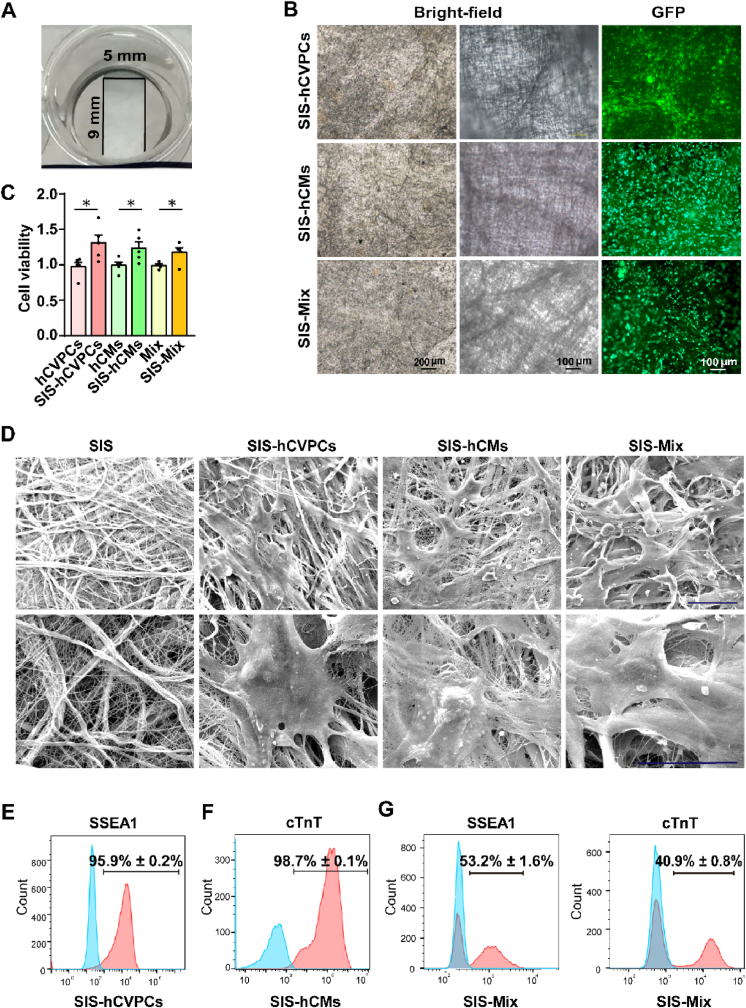
Fig. 1.
Generation and characterization of hiPSC-derived cardiac lineage cell-seeding SIS patches. (A) Photographic image of the small intestinal submucosa (SIS) patch in a culture dish. (B) Images of hCVPCs, hCMs, and the Mix (hCVPCs: hCMs, 1:1) seeding on the SIS patch at a cell density of 3 × 105/cm2 obtained under a bright-field (left panel, at low magnification, middle panel, at high magnification) and fluorescence (right panel) microscopy. (C) Analyses of cell viability of hCVPCs, hCMs, and Mix cultivated on the SIS patches or without the patches by using the Cell Counting Kit-8 (CCK-8) assay. n = 5. Data are mean ± SEM. Statistical significance was assessed using t-test. *p < 0.05. (D) Images of the SIS, SIS-hCVPCs, SIS-hCMs, and SIS-Mix obtained from scanning electron microscope. Scar bars, 50 μm. (E–G) Flow cytometry analysis of SSEA1+ (hCVPC marker) and cTnT+ (cardiomyocyte marker) populations of the hCVPCs (E), hCMs (F), and Mix (G) seeded on the SIS patches. The blue filled histogram, isotype control; and the red filled histogram, the positive population. n = 3 each.