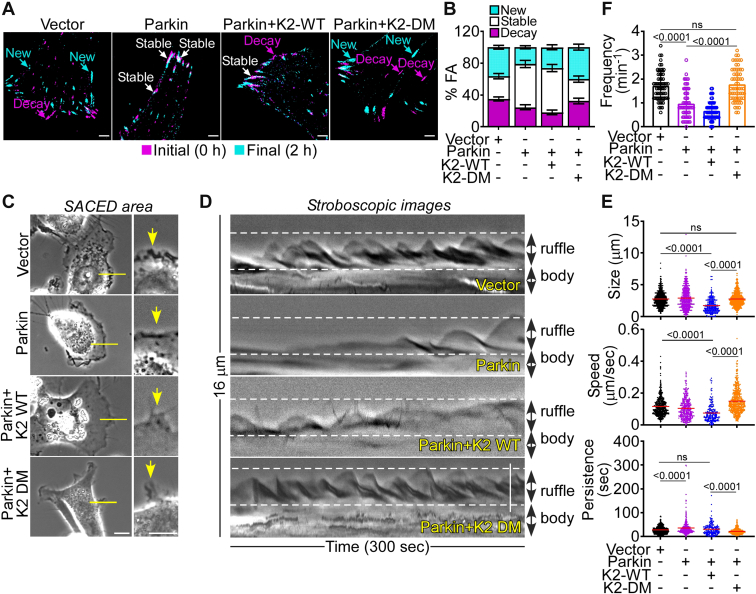
Figure 2.
Modulation of plasma membrane dynamics.A and B, Parkin-expressing LN229 cells were reconstituted with K2-WT or K2-DM, labeled with Talin-GFP and imaged by time-lapse videomicroscopy (A) with quantification of stable, decay, and new FA (B). Representative merged frames at 0 h (magenta) and 2 h (cyan) are shown. The scale bar represents 15 μm. Mean ± SEM (n = 17–19). The statistics are as follows: new FA, vector vs. Parkin, p = 0.009; vector versus Parkin + K2-WT, ns; vector versus Parkin + K2-DM, ns; stable FA, vector versus Parkin, p < 0.0001; vector versus Parkin + K2-WT, p < 0.0001; vector versus Parkin + K2-DM, ns; decay FA, vector versus Parkin, ns; vector versus Parkin + K2-WT, p = 0.003; vector versus Parkin + K2-DM, ns. C, LN229 cells as in (A) were analyzed by real-time phase-contrast imaging. Representative images show individual cells and one area examined for membrane dynamics (arrows). The scale bar represents 10 μm. D, cell membrane dynamics for each discrete area in (C) is represented as a stroboscopic image, where the cell body and ruffle/lamellae are labeled. The scale bar represents 10 μm. E, each individual cell protrusion was analyzed for lamella size (top), speed of retraction (middle), and lamella persistence (bottom). Each point represents an individual membrane protrusion with 203 (Parkin + K2-WT), 299 (Parkin), 513 (vector), and 532 (Parkin + K2-DM) membrane events from 15 individual cells per group. Mean values per group are indicated. F, the conditions are as in (E), and the frequency of lamella dynamics was quantified. Mean ± SD (n = 60). For all panels, numbers correspond to p values by one-way ANOVA with Tukey multiple comparisons test. ns, not significant.