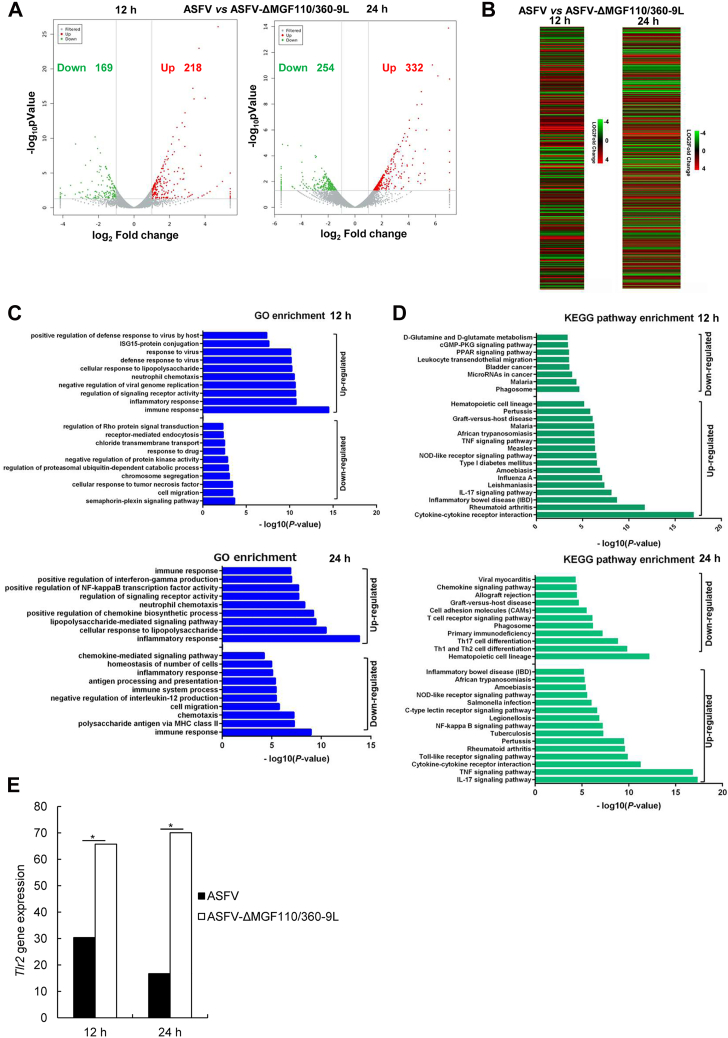
Figure 5.
Transcriptome profiles of primary porcine alveolar macrophages infected with ASFV CN/GS/2018 or ASFV-ΔMGF110/360-9L (MOI = 0.1) at 12 or 24 h.A, volcano map of differentially expressed genes (DEGs) in PAMs infected with ASFV CN/GS/2018 or ASFV-ΔMGF110/360-9L. Red dots indicate significantly upregulated DEGs, green dots indicate significantly downregulated DEGs, and gray dots indicate no significant change. B, heat map of DEGs in PAMs infected with ASFV CN/GS/2018 or ASFV-ΔMGF110/360-9L. C, GO terms at 12 h (top panel) and 24 h (bottom panel). D, KEGG pathway enrichment at 12 h (top panel) and 24 h (bottom panel). E, Tlr2 gene expression of PAMs infected with ASFV CN/GS/2018 (filled bar) or ASFV-ΔMGF110/360-9L (unfilled bar). The graphs show the mean ± SD. In the displayed experiment, three replicates were included per experimental condition. ∗p < 0.05, represent the ASFV and ASFV-ΔMGF110/360-9L groups. ASFV, African swine fever virus; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PAM, porcine alveolar macrophage; MOI, multiplicity of infection; TLR2, Toll-like receptor 2.