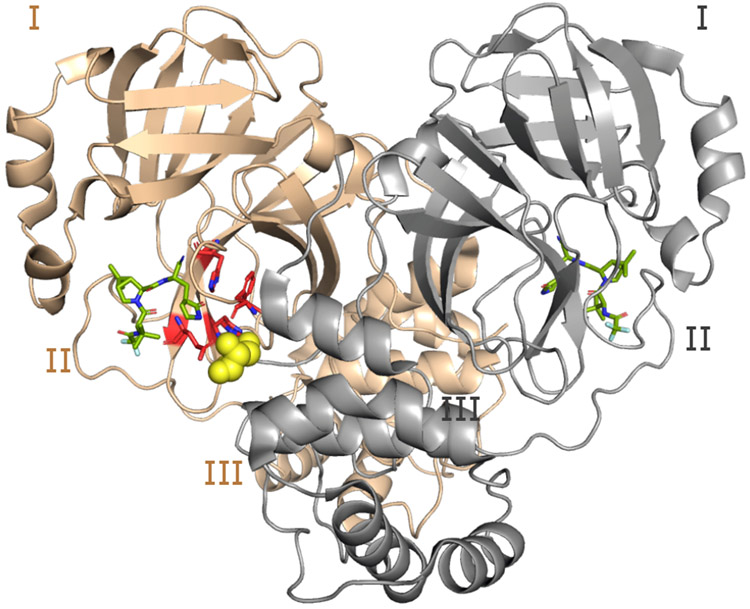
Figure 1: Structure of the WT SARS-CoV-2 Mpro dimer.
Cartoon representation of the Mpro dimer bound to nirmatrelvir (PDB ID 7vh85) with protomer A in tan and B (front of the image) in grey. The three domains (I, II, and III) are labeled for each protomer. The S1 pocket residues (Phe140, His163, Glu166, and His172) of protomer A (highlighted in red and shown as sticks) interacts with Ser1* from protomer B (shown in the van der Waals sphere representation). Ser1* forms either a hydrogen bond or salt bridge with Phe140, Glu166, and His173, while His163 forms aromatic stacking with Phe140. The inhibitor nirmatrelvir is shown in green.