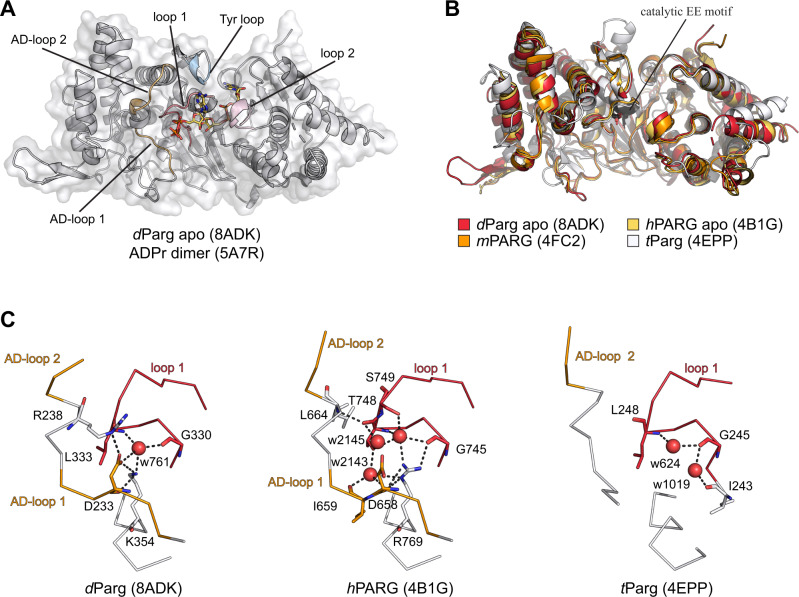
Fig. 10. Structural basis of mono-Ser-ADPr hydrolysis by dParg.
A Ribbon-surface representation of apo dParg. ADP-ribose dimer (yellow) of the aligned hPARG:ADP-ribose dimer complex structure is given to highlight the active site. Structural features important for catalysis are highlighted: accessory-domain loops 1 and 2, AD-loop 1 and 2; catalytic loop, loop 1; diphosphate binding loop, loop 2; tyrosine clasp, Tyr loop. B Ribbon representation of structural alignment of PARG domains from indicated species (Drosophila, red; human, yellow; mouse, orange; T. thermophile, white). The catalytic residues (Glu340/Glu341 in dParg) are highlighted in black. C Ribbon-liquorice representation of loop 1-AD-loop 1 interaction of Drosophila, human, and T. thermophile PARGs. Polar interactions are highlighted as black dotted lines and water molecules involved in the interaction as red spheres.