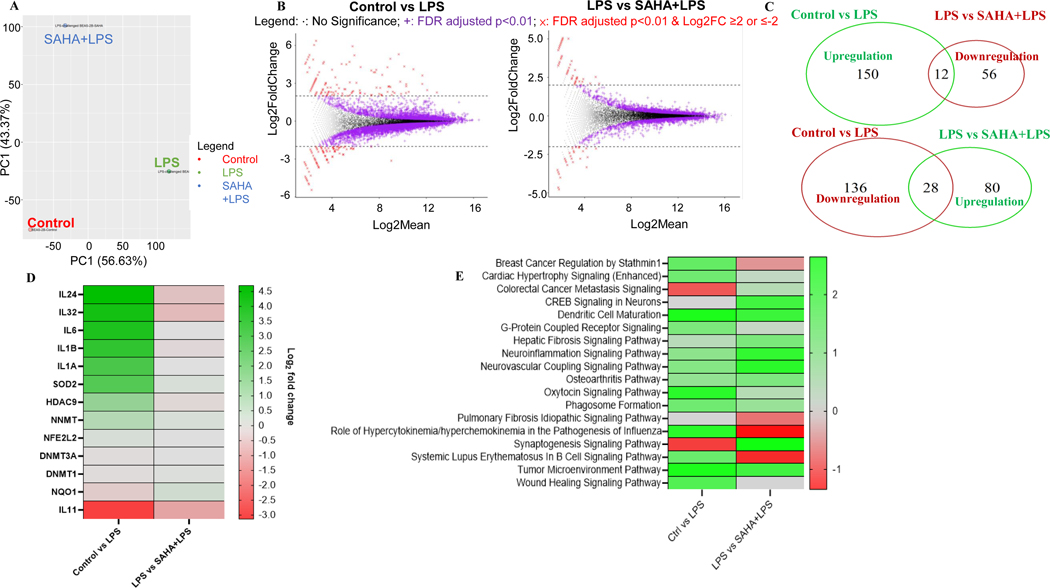
Figure 5. Transcriptomic changes by suberoylanilide hydroxamic acid (SAHA) in lipopolysaccharide (LPS)-challenged BEAS-2B cells.
(A) PCA plot transcriptomic profiles and Dendrogram of the gene expression profiles clustered by Euclidean distance; (B) MA plots showing DEGs in response to LPS vs. Control group and LPS+SAHA vs. LPS group with a cut-off of p<0.01 and log2(fold change) ≥ 2 or ≤ −2; (C) Venn diagrams comparing the upregulated and downregulated genes in LPS vs. Control group and LPS+SAHA vs. LPS group; (D) Circos plot of top 11 DEGs that appeared in the completely opposite direction in LPS vs. Control group and LPS+SAHA vs. LPS group; (E) Signaling pathway analysis of SAHA by IPA. RNA samples in all groups (n = 1) were pooled from three replicates.