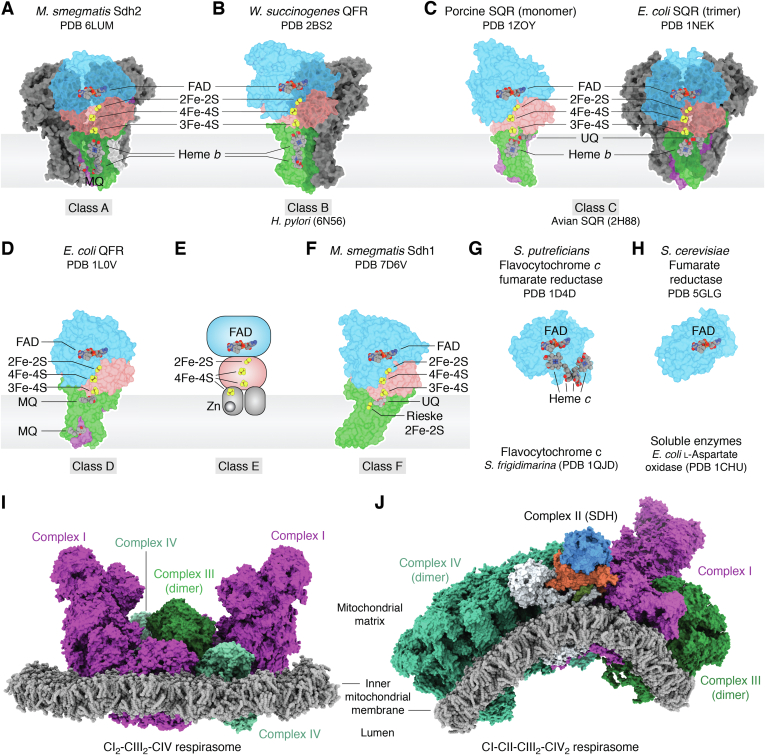
Figure 4.
Diversity in structure and megastructure across the greater complex II family. Complex II enzymes are currently categorized into six recognized classes based on their membrane-spanning regions (83). Most members of the complex II superfamily contain a soluble domain with two subunits and between one and three membrane-spanning subunits, although Class B members are believed to peripheral-membrane enzymes and there are fully soluble homologs. While all complex II homologs share significant sequence identity in the soluble domains, the membrane-spanning domains may have evolved independently, possibly more than one time. In addition, some complex II family members contain supernumerary subunits. Depending on whether they were discovered for a physiological role in aerobic or anaerobic respiration, the corresponding complex II genes are termed sdhABCD (aerobic form) or frdABCD (anaerobic form). A, Class A complex II enzymes are predominantly from archaea. These can contain three membrane-spanning subunits, two of which have three membrane-spanning helices, with the additional single-pass membrane-spanning subunit termed SdhF. Class A homologs house two integral-membrane b-type hemes. Shown is the cryoEM structure of Mycobacterium smegmatis Sdh2, which is a trimer (PDB 6LUM (77)). B, Class B complex II contains a single membrane-spanning subunit with five membrane-spanning helices and two b-type hemes. Class B complex II enzymes are fumarate reductases and obligate dimers. Shown is the X-ray crystal W. succinogenes QFR (PDB 2BS2 (59)). C, Class C complex II enzymes contain two membrane-spanning helices and one b-type heme. Members of Class C complex II are of intense interest because they include mitochondrial homologs. Some Class C enzymes are (left) monomers (porcine mitochondrial complex II, PDB 1ZOY (75)) while others are (right) trimers (E coli SQR, PDB 1NEK (42)). D, Class D complex II enzymes are monomers that contain two membrane-spanning helices but no integral membrane heme. A putative cofactor is located at a similar position as heme in other homologs, but the identity of this has not been assigned (223). Shown is the E. coli QFR (PDB 1L0V (58)). E, Class E complex II enzymes contain membrane anchors with an amphipathic nature that suggests they could be peripherally associated with the membrane (224). Class E complex II enzymes coordinate a Zn2+ and 4Fe-4S cluster in the amphipathic helices. A structure representing this class has not yet been reported. F, Class F complex II enzymes contain a single membrane-spanning subunit with six helices. Members of Class F coordinate an integral-membrane Rieske Fe-S cluster. Shown is the structure of Mycobacterium smegmatis Sdh1 (PDB 7D6V (79)). G and H, soluble homologs of the SDHA subunit have distinct biological roles. G, some soluble SDHA homologs contain a fused tetra-heme domain and biologically act as fumarate reductases in anaerobic respiration (60, 61, 62). Shown is the Shewanella oneidensis (formerly putrefaciens) flavocytochrome c fumarate reductase (PDB 1D4D (62)). H, some soluble SDHA homologs are found without additional fused polypeptide and can catalyze redox reactions related to dicarboxylate oxidoreduction. The most extensively studied soluble SDHA homolog is perhaps L-Aspartate oxidase. Shown is the yeast Osm1 fumarate reductase (PDB 5GLG (94)). I and J, numerous structures of respiratory supercomplexes have been reported, with varying stoichiometries. These all contain complex III (CIII) at the core, which is an obligate dimer. Complex I (CI) and complex IV (CIV) can each interact with CIII independently and have one or two copies associated with each CIII2. I, reported respirasomes lacking complex II include: CICIII2, CIII2CIV, CICIII2CIV, and CI2CIII2CIV2. Shown is the human CI2CIII2CIV2 respirasome (PDB 5XTI, (110)) with CI in magenta, CIII in green, and CIV in teal. J, the only reported structure of a respirasome that contains complex II (CII) is from the ciliate Tetrahymena thermophila and has a stoichiometry of CICIICIII2CIV2 (80). Note that Tetrahymena respiratory complexes contain supernumerary subunits as compared to their counterparts in animals (111). This is particularly notable in CIV and in CII, with the latter containing 11 supernumerary subunits (SDHTT1 – SDTT11). For complex II, SDHA is in blue, SDHB is in red, SDHC and SDHD are in green and purple, and the 11 supernumerary subunits are in white. SDH, succinate dehydrogenase; QFR, quinol:fumarate reductase.