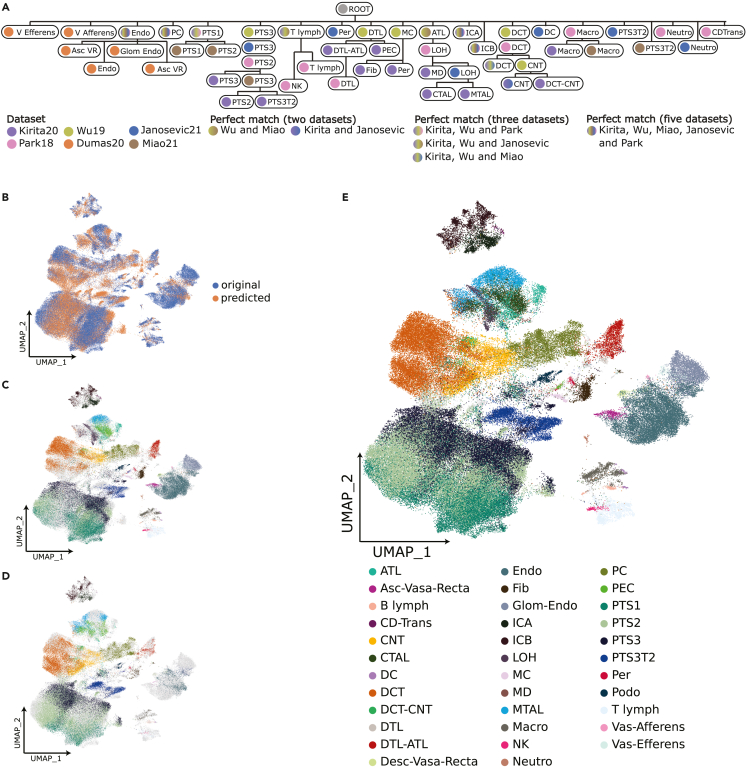
Figure 3.
Hierarchically defined kidney mouse atlas
(A) Learned classification tree on the four annotated datasets after manually harmonizing annotations. Tree nodes are colored by the supporting dataset or datasets in case of two or more cell populations matching.
(B) UMAP plot visualizing cells used to train the classification tree (blue) and cells for which cell type was not known (orange).
(C) UMAP representation of annotated cell typesPark18, Kirita21, Dumas20, Miao21, Janosevic21, and Wu19.
(E) UMAP embedding of predicted cell types for Hinze20, Conway20 and cells labeled “unknown” or “missing.” e UMAP plot combining predicted and available annotations resulting in the integrated mouse kidney atlas. IC: Intercalated Cell, ICA: Intercalated Cell Type A, ICB: Intercalated Cell Type B, Endo: Endothelial Cell, Fib: Fibroblast, Macro: Macrophage, B lymph: B lymphocyte, Stroma: Stroma cell, NK: Natural Killer, T lymph: T lymphocyte, PT: Proximal Tubule, PTS1: Proximal Tubule Segment 1, PTS2: Proximal Tubule Segment 2, PTS3: Proximal Tubule Segment 3, PTS3T2: Proximal Tubule Segment 3 Type 2, PC: Principal Cell, PEC: Parietal Epithelial Cell, Per: Pericyte, DCT: Distal Convoluted Tubule, ATL: Ascending Thin Limb of Henle, MD: Macula Densa, LOH: Loop of Henle, CTAL: Thick Ascending Limb of Henle in Cortex, MTAL: Thick Ascending Limb of Henle in Medulla, CNT: Connecting Tubule, Podo: Podocyte, DTL: Descending Thin Limb of Henle, MC: Mesangial Cell, Neutro: Neutrophil, Asc-Vas-Recta (Asc VR): Ascending Vasa Recta, Desc-Vas-Recta (Desc VR): Descending Vasa Recta, Glom Endo: Glomeruli Endothelial, V afferens: Vas Afferens, V Efferens: Vas Efferens.