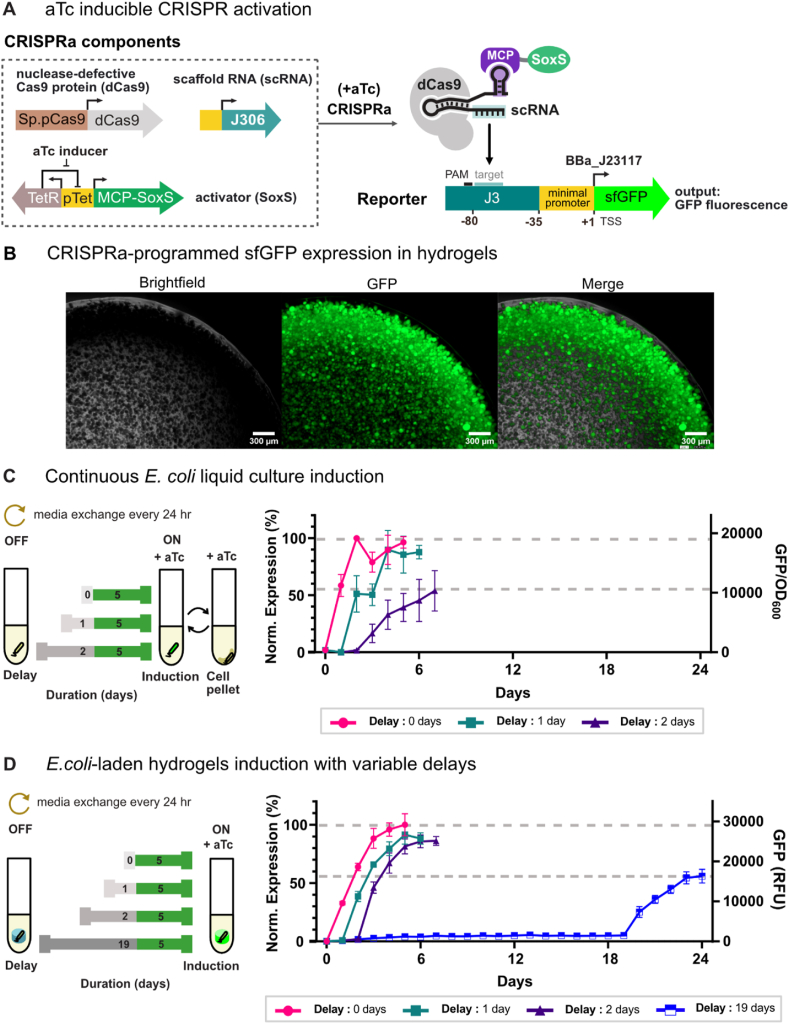
Fig. 2.
Persistence of genetic activity in E. coli ELMs. a) Schematic of an inducible CRISPR activation (CRISPRa) program for sfGFP expression. CRISPRa-directed expression of sfGFP uses nuclease-defective Cas 9 (dCas9) and a scaffold RNA (scRNA) that specifies a target site upstream of the engineered J3 promoter. scRNA (J306) is a modified guide RNA that includes a 3′ MS2 hairpin to recruit a transcriptional activator (SoxS) fused to the MS2 coat protein (MCP). The CRISPRa-programmed expression of sfGFP is induced upon the addition of anhydrotetracycline (aTc), which expresses MCP-SoxS from the pTet promoter. b) CRISPRa-programmed sfGFP expression in hydrogel-encapsulated E. coli. Microscopy images were taken from a two-day old continuously-cultured ELM (scale bar = 300 μm). c) Gene expression dynamics were evaluated in bacterial liquid continuous culture following variable delays of 0, 1, and 2 days (gray) before CRISPRa-programmed sfGFP expression was initiated and carried out for 5 days (green) by the addition of aTc inducer. Media was continuously removed and replenished every 24 h throughout the entire experiment, keeping the culture volume and composition constant via cell pelleting. sfGFP expression was tracked over time following continuous culture induction delays. Values represent the mean ± standard deviation from n = 3 replicates. d) Gene expression dynamics in ELMs were evaluated following variable delays of 0, 1, 2 and 19 days (gray) of continuous culture before initiation of CRISPRa-programmed sfGFP expression for 5 days (green). Media was replenished every 24 h throughout the entire experiment. The impact of induction delay duration on ELM gene expression levels was quantified by tracking sfGFP expression. Values represent the mean ± standard deviation from n = 5 technical replicates, except for 19-day delay samples, where n = 3 technical replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)