Figure 1. HPV multimers in cancer cell lines.
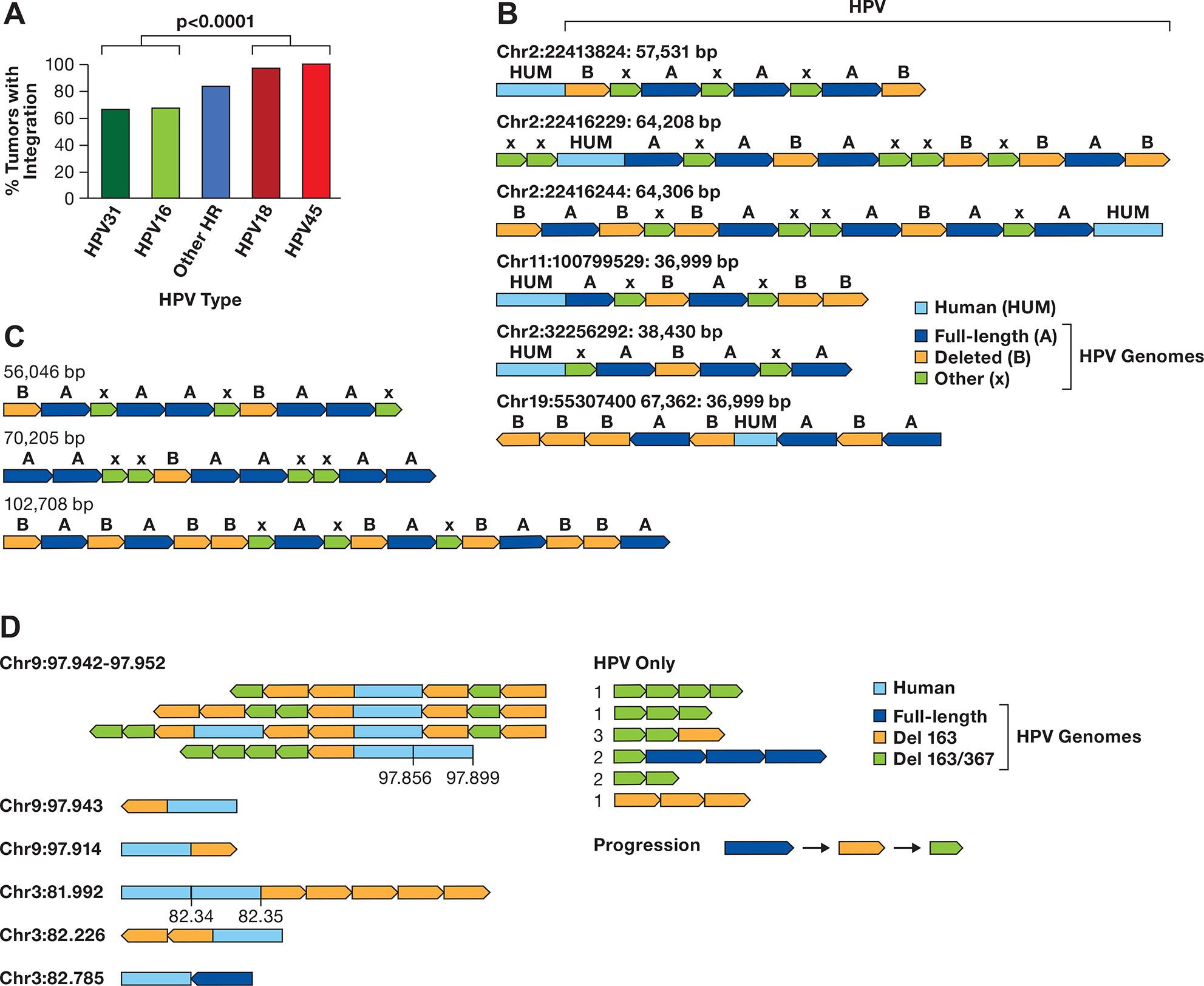
A. The integration frequency of the predominant carcinogenic HPV types is shown using combined data from TCGA and Guatemalan tumors (4,27). The other high-risk (HR) types are combined. The number of samples is HPV31 (N=3), HPV16 (N=170), Other HR (N=43) includes 13 HPV types, HPV18 (N=32), and HPV45 (N=17). The comparision of HPV16/31 to HPV16/45 is significant, P> 0.0001 byt Fishers exact test. B. A diagram of contigs of long DNA sequence reads in CaSki cells aligning to the human genome is shown. Human DNA sequences are in light blue, the full-length HPV16 genome A is dark blue, a 6.5 kb deleted genome derived from genome A is orange, and other smaller HPV16 fragments of varying size are green (X). The arrows show the direction of the HPV genome segments. The location of the human junction and size of the contig is shown. C. HPV only reads from CaSki cells, key as in panel B. D. Contigs of SCC152 HPV16 multimers are shown, with the human/HPV junction location on the left in Mb. For reads with an additional human segment, those coordinates are given above the contig. For HPV-only reads, the number of times that structure was found are shown to the left of the read diagram. A suggested progression in the evolution of the single and double deleted HPV16 genomes is shown.
