Figure 5. Diagram displaying data from 62 tumors.
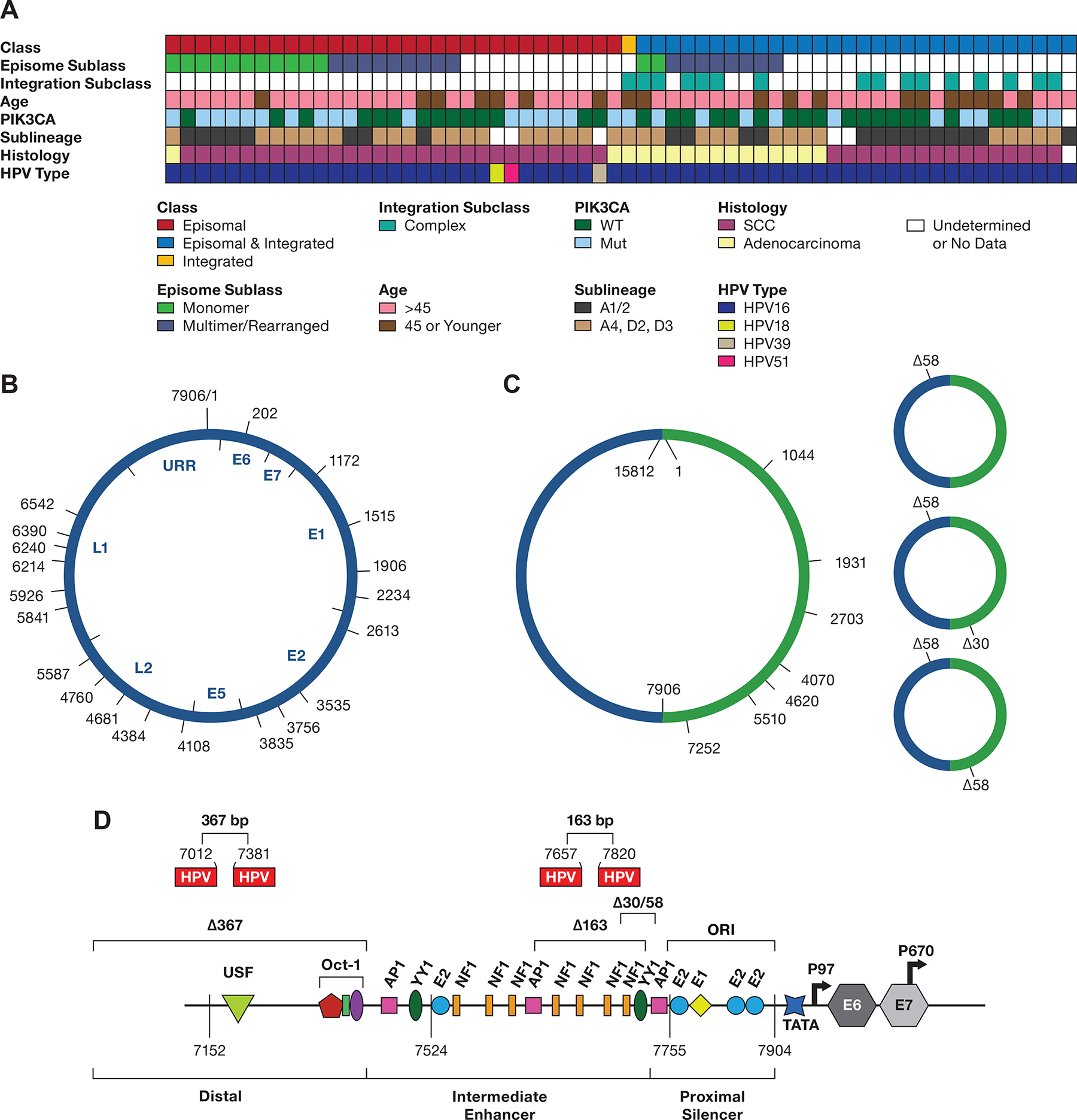
A. The integration class and subclass, episomal subclass, age range, PIK3CA mutation status, HPV type and HPV16 sublineage, and histology are shown. SCC, squamous cell carcinoma; WT, wild type; Int, integration. Blanks represent samples not able to be classified. B. Diagram of the 7906 bp HPV16 genome DNA, displaying the start site of DNA sequence reads from 13 tumors containing a complete or nearly complete monomer genome sequence. The position of the HPV16 genes and URR are shown inside the diagram. C, A diagram of HPV16 dimers from three tumors showing the start position of reads with complete or nearly complete copies of two tandem HPV16 genomes (Supplemental Table 10). D, the location of 30 and 58 bp deletions in the URR occurring in dimer reads from tumor T393.The location of the 163 and 367 bp deletions in the SCC152 cell line is shown, as well as their locations in the HPV16 upstream regulatory region. The 367 bp deletion removes most of the distal region, and the 163 bp deletion removes four NF1 binding sites and two YY1 binding sites (only one is shown) in the Intermediate Enhancer region. Shown are the binding sites for the viral E1 and E2 proteins and the transcription factors OCT1, AP1, YY1, and NF1. ORI, the origin of replication; TATA, TATA-binding site; P97, major promoter; P670, minor promoter. The locations of the coding region of the E6 and E7 genes are indicated
