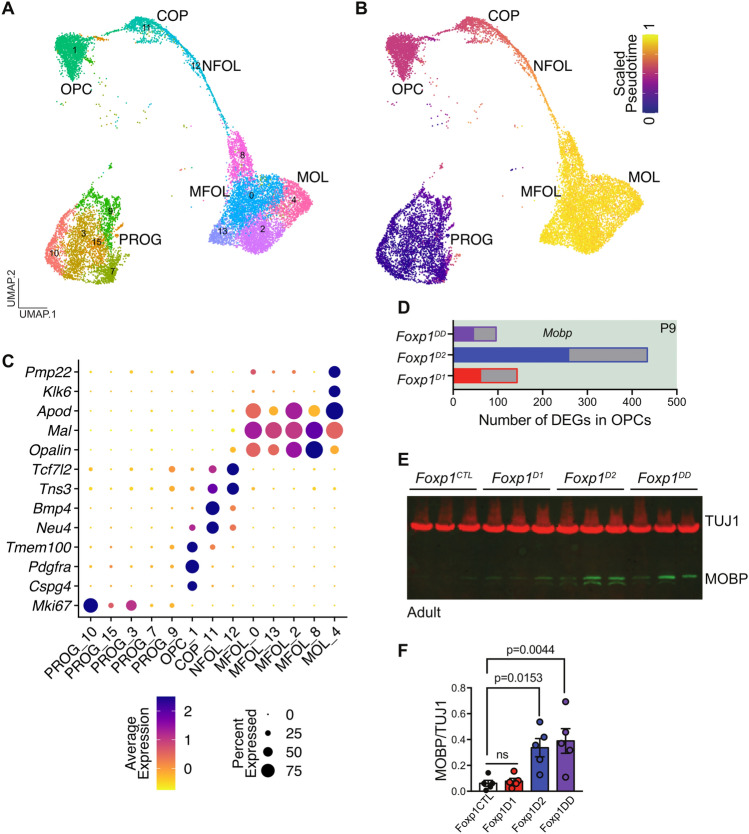
Figure 5.
Pseudotime analysis of oligodendrocytes across striatal development and with deletion of Foxp1. (A) Annotated UMAP of oligodendrocyte progenitor cells (OPCs), committed oligodendrocyte precursors (COPs), newly-formed oligodendrocytes (NFOL), myelin-forming oligodendrocytes (MFOL), mature oligodendrocytes (MOL) and progenitor clusters from the combined dataset. (B) Cells colored by PHATE pseudotime values. (C) Scaled expression of genes specific to each oligodendrocyte subtype. (D) Number of differentially expressed genes (DEGs) in OPCs from P9 dataset with deletion of Foxp1 from dSPNs, iSPNs, or both. Filled bars are upregulated DEGs (such as Mobp) and grey bars are downregulated DEGs. (E) Western blot of TUJ1 (housekeeping gene) and MOBP (oligodendrocyte marker) in striatal tissue in Foxp1CTL, Foxp1D1, Foxp1D2, and Foxp1DD mice and (F) quantification of MOBP levels relative to TUJ1 across genotypes (N = 5/genotype). Data is represented as mean ± SEM. P-values determined using a one-way ANOVA with Dunnett’s multiple comparison.