Fig. 1. CRISPR-based technologies for small and large genome edits.
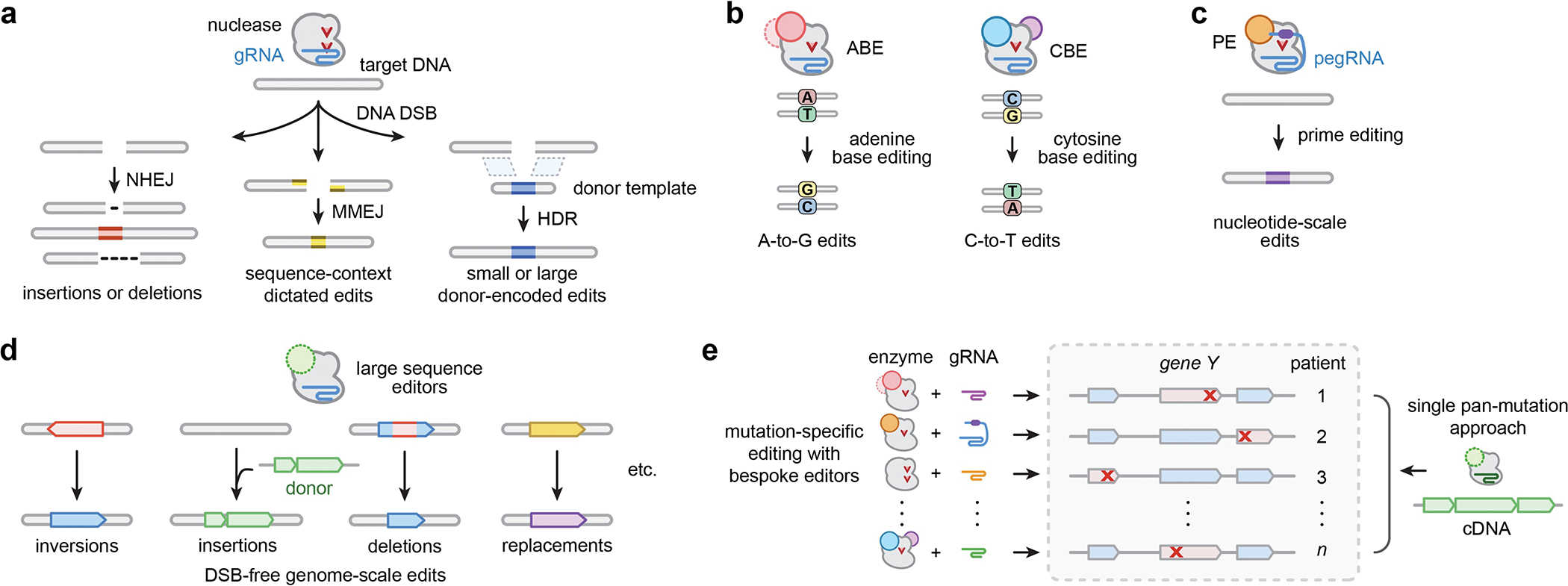
a, Summary of major edit outcomes following nuclease-mediated editing. CRISPR-Cas enzymes paired with guide RNAs (gRNAs) generate DNA double-strand breaks (DSBs) to initiate editing events, which are generally repaired by non-homologous end joining (NHEJ), micro-homology-mediated end joining (MMEJ), or homology-directed repair (HDR). b, Adenine and cytosine base editors (ABEs and CBEs) generate A-to-G and C-to-T changes, respectively, without intentionally causing DSBs or requiring HDR. c, Prime editors (PEs) generate short insertion, deletion, and/or substitution edits that are encoded on a prime editing guide RNA (pegRNA). d, Large sequence editors can generate targeted inversions, insertions, and deletions of multi-kb sequences, which would enable novel editing approaches for previously inaccessible classes of diseases. e, Compared to the requisite suite of mutation-specific gene editing approaches for diseases caused by numerous heterogeneous mutations, multi-kilobase integration and replacement technologies could act as single pan-mutation therapies for all genotypes.
