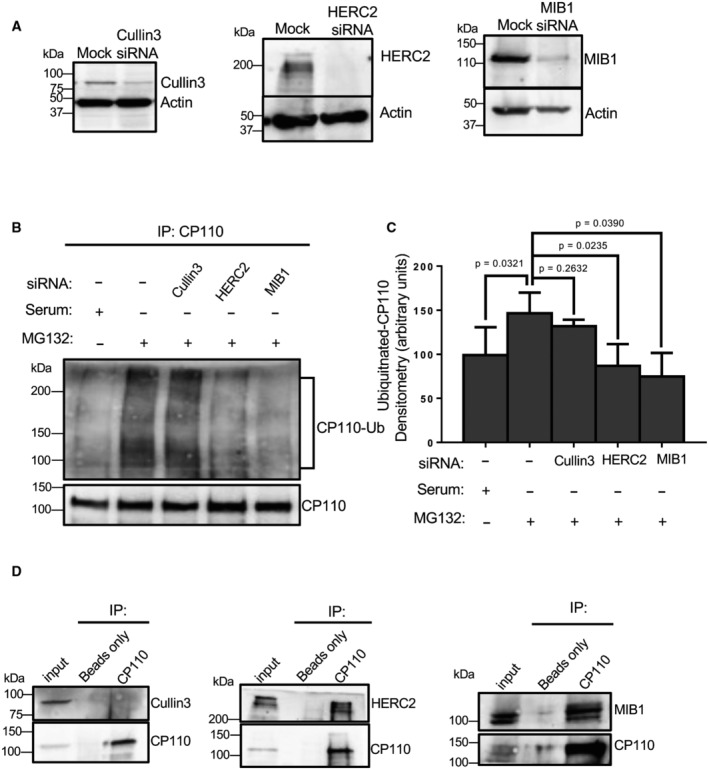
Figure 3. Depletion of HERC2 or MIB1 impedes CP110 ubiquitination upon serum starvation.
-
AValidation of siRNA depletion. RPE‐1 cells were mock‐transfected or transfected with siRNA oligonucleotides to impair translation of Cullin3, HERC2, or MIB1. 72 h later the cells were lysed, subjected to SDS–PAGE and immunoblotted. Immunoblotting with anti‐actin antibodies was done as a loading control.
-
BHERC2 or MIB1 depletion impairs CP110 ubiquitination. RPE‐1 cells were grown in serum (first lane from left), or with MG132 in the absence of serum for 4 h (second lane from left), transfected with Cullin3 siRNA oligonucleotides and treated with MG132 in the absence of serum for 4 h (third lane from left), transfected with HERC2 siRNA oligonucleotides and treated with MG132 in the absence of serum for 4 h (fourth lane from left), or transfected with MIB1 siRNA oligonucleotides and treated with MG132 in the absence of serum for 4 h (fifth lane from left). Cells were lysed and immunoprecipitated with anti‐CP110 antibodies, and the resultant immunoprecipitates were immunoblotted with anti‐ubiquitin to identify ubiquitinated CP110 (upper panel). The blot was then stripped and reimmunoblotted with anti‐CP110 to reveal the amount of CP110 pulled down from the lysate (lower panel).
-
CGraph displaying densitometric analysis of ubiquitinated CP110 bands normalized to total CP110 immunoprecipitated. Normal distribution was determined by the Shapiro–Wilk normality test, and statistical significance was calculated with an unpaired two‐tailed t‐test.
-
DCP110 interacts with HERC2 and MIB1, but not Cullin3. RPE‐1 cells were lysed and immunoprecipitated with beads only or anti‐CP110, and then subjected to SDS–PAGE and immunoblotting with CP110 to validate the immunoprecipitation (lower panel of each gel) and either Cullin3 (left gel, upper panel), HERC2 (middle gel, upper panel) or MIB1 (right gel, upper panel). 7% of inputs are shown on the left of each gel.
Data information: All gels are representative ones from three individual biological experiments. Graph displays mean densitometry representing ubiquitination of CP110 from three individual experiments, bars indicate standard deviation, and statistical significance was calculated with an unpaired two‐tailed t‐test, since the Shapiro–Wilk test indicated a normal distribution.
Source data are available online for this figure.
