Figure EV1. The estimation of “Dex‐trade‐off” adaptive mutations.
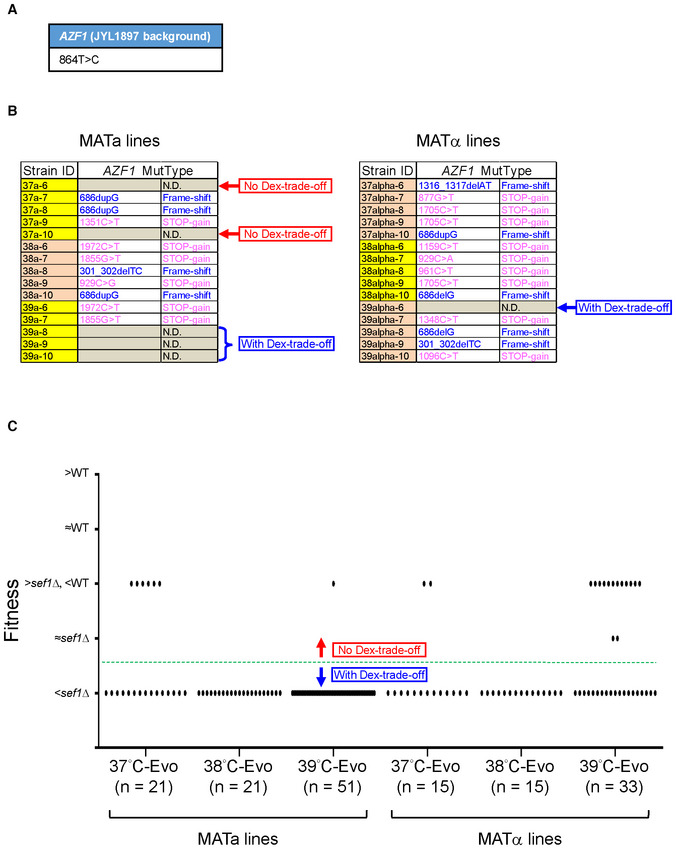
- The parental SNP polymorphism of L. kluyveri AZF1 in the JYL1897 background (the wild‐type of our lab) in comparison with the published reference genome of CBS 3082.
- The AZF1 mutational spectrum of 30 more randomly picked heat stress‐evolved sef1Δ suppressors (five clones each from 37, 38, and 39°C‐Evo sef1Δ suppressors in both MATa and MATα lineages). The mutations were identified by Sanger sequencing. “N.D.” means that there is no mutation detected within the coding region. Whether the clones that carry no azf1 mutation displayed “Dex‐trade‐off” phenotypes or not is indicated on the right side of each table.
- The “Dex‐trade‐off” phenotypic spectrum of 156 heat stress‐evolved sef1Δ suppressors. The “Dex‐trade‐off” phenotypes of each clone are displayed according to the simple fitness scores under the YPD_37°C condition (Dataset EV3). Clones with score 1 (with fitness worse than sef1Δ strain) were defined as “with Dex‐trade‐off” (please also see Appendix Fig S3B for the simple fitness score definition). About 14.74% (23/156) of clones in this batch showed no clear “Dex‐trade‐off.”
