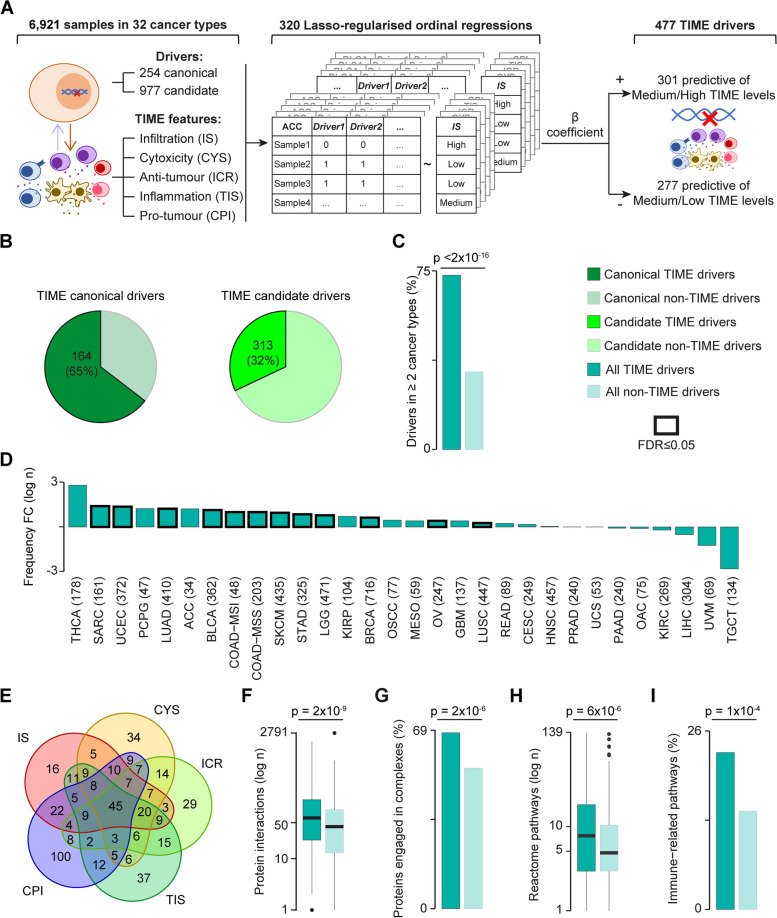
Fig. 1.
Identification and properties of TIME drivers. A Approach for TIME driver identification. Interactions between damaging alterations in cancer-specific drivers and TIME features were assessed in 32 TCGA cancer types using Lasso-regularised ordinal regression. Regressions were computed separately for canonical and candidate drivers and TIME features in each cancer type. 0 and 0 indicated that altered cancer drivers were predictive of medium/high or medium/low TIME levels, respectively. B Proportions of TIME canonical and candidate drivers over all drivers. C Proportions of TIME and non-TIME drivers in ≥ 2 cancer types. D Fold change (FC) of median frequency alterations of TIME and non-TIME drivers in each cancer type. The number of samples in each cancer type is shown in brackets. E Venn diagram of TIME drivers predictive of the five TIME features. F Distributions of protein–protein interactions in TIME and non-TIME drivers. G Proportions of TIME and non-TIME drivers that are part of protein complexes. H Distributions of level 2–9 Reactome pathways [39] containing TIME and non-TIME drivers. I Proportion of TIME and non-TIME drivers mapping to immune-related pathways as derived from MSigDB [40] and Reactome [39]. CPI, cancer-promoting inflammation; CYS, cytotoxicity score; FDR, false discovery rate; ICR, immunologic constant of rejection; IS, immune score; TIS, tumour inflammation signature. TCGA abbreviations are listed in Additional File 2: Table S1. Proportions (C, G, I) and distributions (D, F, H) were compared using Fisher’s exact test and Wilcoxon rank-sum test, respectively. In D, Benjamini–Hochberg correction for multiple testing was applied