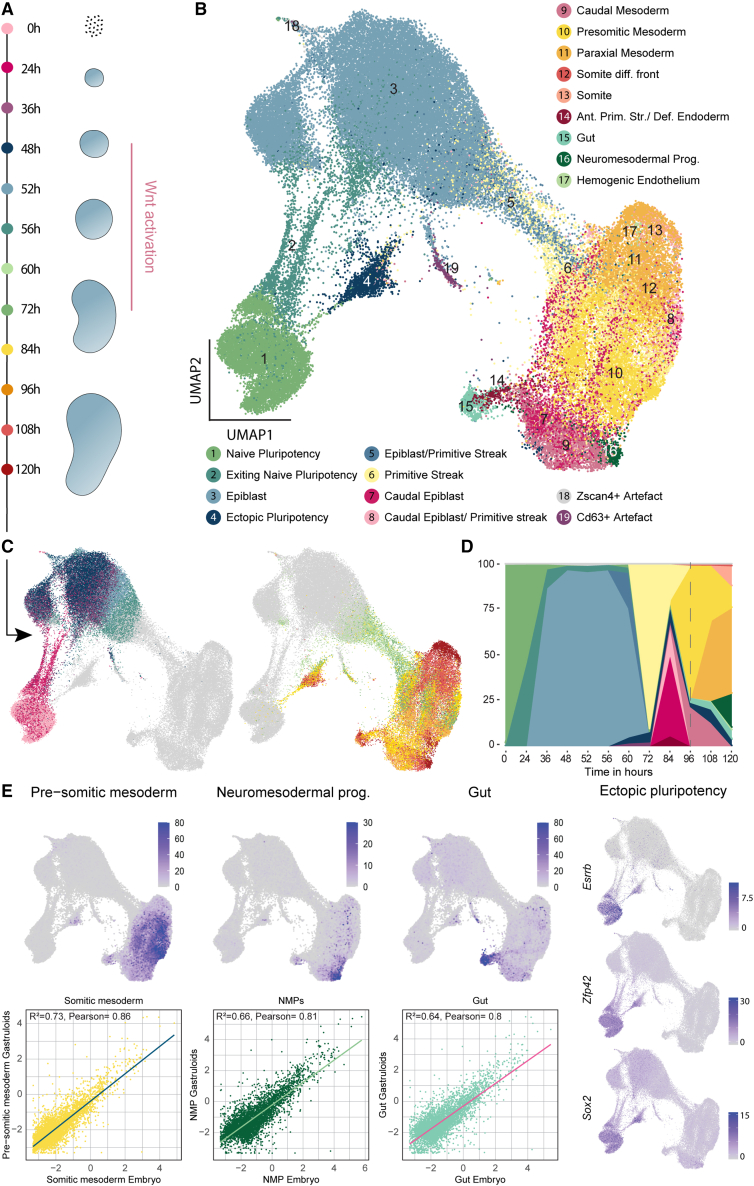
Figure 1.
scRNA-seq time course of gastruloid development
(A) Scheme of gastruloid formation between 0 and 120 h, including sampling time points for scRNA-seq. Pink bar: Wnt activation.
(B) Uniform Manifold Approximation and Projection (UMAP) of scRNA-seq time course experiments including cell type annotations. Somite diff. front, somite differentiation front; Ant. Prim. Str., anterior primitive streak; Def. endoderm, definitive endoderm; NMPs, neuro-mesodermal progenitors.
(C) UMAP of single-cell transcriptomes highlighting sampling time points. Legend: (A).
(D) Ribbon plot showing changes in cell type composition over time. Dashed line: 96 h.
(E) UMAPs showing the aggregated expression of marker genes for the NMPs (Hes3, Hoxb9, Cdx4, and Epha5), gut (Sox17, Foxa2, Cer1, Krt8, Krt18, Shh, and Gsc), and pre-somitic mesoderm (Hes7, Aldh1a2, Dll1, Tbx6, Cyp26a1, and Hoxb1) (top). Scatter plots and inferred linear regression for conservation analysis between gastruloid and embryonic cells. Scatter plot: log2 mean expression of genes in gastruloid and embryonic cell types. Expression UMAPs for pluripotency genes Esrrb, Zfp42, and Sox2 (right).