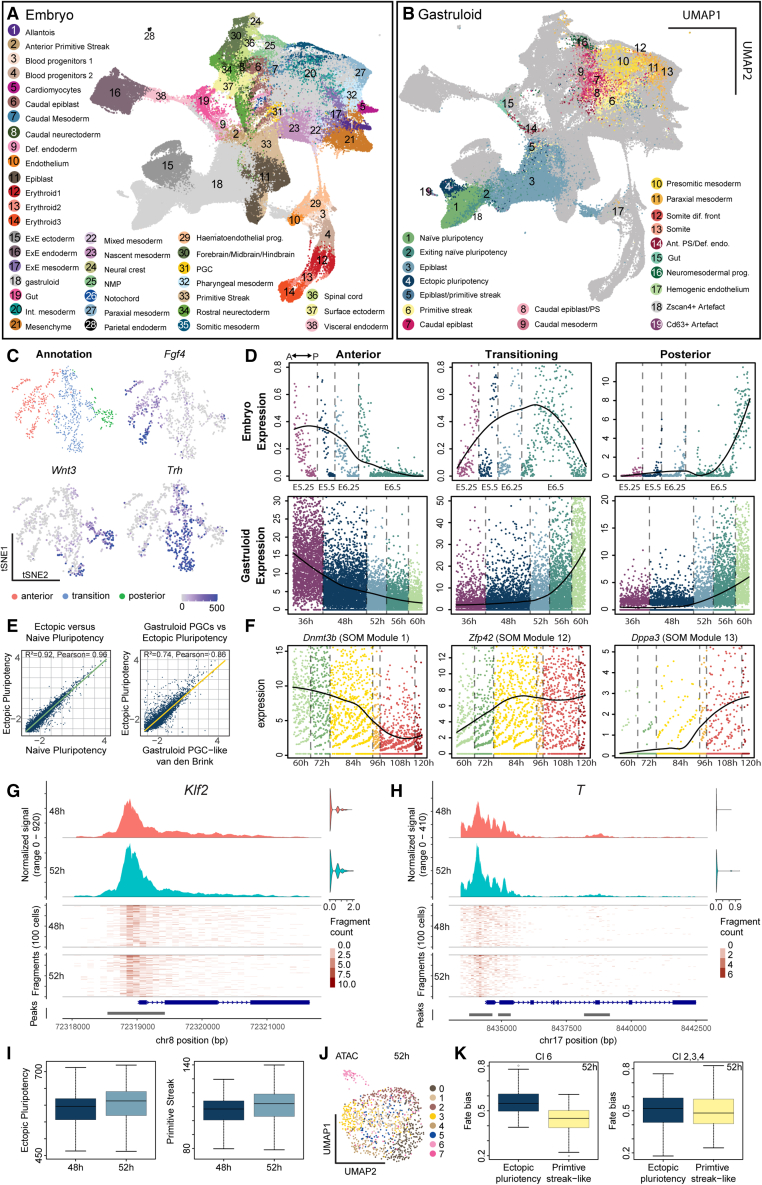
Figure 2.
In vivo comparison and characterization of epiblast and pluripotency states
(A) UMAP of co-embedded gastruloid and embryonic cells highlighting embryonic cell types from Pijuan-Sala et al.23
(B) UMAP of co-embedded gastruloid and embryonic cells highlighting gastruloid cell types. PS, primitive streak; dif., differentiation; Ant. PS/Def. endo., anterior primitive streak/definitive endoderm.
(C) t-SNE map of single-cell transcriptomes from Cheng et al.27 highlighting three embryonic epiblast states and expression maps of Fgf4, Trh, and Wnt3.
(D) Temporal gene expression maps of anterior, transition, and posterior gene signatures for embryonic and gastruloid epiblast cells. y axis: normalized expression.
(E) Scatter plot and inferred linear regression comparing ectopic and naive pluripotency populations (left) and PGC-like15 populations (right). Scatter plot: mean expression of individual genes in ectopic and naive pluripotency populations or PGC-like populations.
(F) Temporal gene expression maps of Dppa3, Zfp42, Dnmt3b, of EP cells. y axis: normalized expression. Self-organizing map (SOM) modules (see Figure S2F).
(G) Coverage plot of chromatin accessibility for Klf2 and 1,000 bp upstream region from transcription start site (TSS) at 48 and 52 h. Right: multiome RNA expression of Klf2 in the same cells. Top: averaged frequency of DNA fragments within the genomic region. Middle: frequency of fragments within the genomic region for single cells. Lower: arrows indicate transcriptional direction. Bottom: peak coordinates within genome region.
(H) Coverage plot of chromatin accessibility for T and 1,000 bp upstream region from the TSS containing the promoter.
(I) Boxplots: aggregated gene activity scores for the EP and primitive streak-like state.
(J) UMAP of the scATAC-seq data-modality from the multiome highlighting clusters. Time point: 52 h.
(K) Boxplots of fate bias analysis at 52 h for multiome clusters toward EP and primitive steak-like populations.