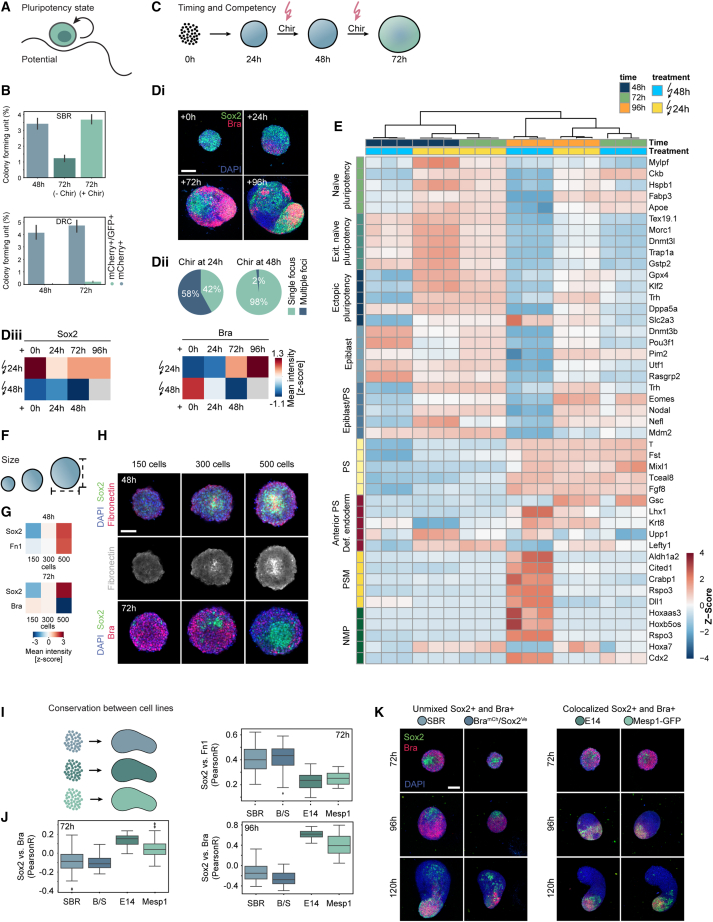
Figure 6.
Core characterization
(A) Scheme illustrating the stem cell pluripotency state.
(B) Top bar plots: colony formation efficiency for SBR gastruloids at 48 and 72 h with or without Wnt activation (−Chir, Chir). Bottom barplots: colony formation efficiency for single cells sorted from DRC gastruloids at 48 and 72 h. Blue bars: clonogenicity of mCherry+. Green bars: efficiency for double-positive cells. Clonogenicity score: fraction of seeded cells forming a colony in %. Error bars show standard deviation for 12 wells/condition.
(C) Scheme showing different Wnt activation timings.
(D) Di: representative images of gastruloid pulsed at different time points. (n = 334 at +0 h, n = 328 at +24 h, n = 299 at +72 h, n = 231 at +96 h.) Middle z plane (+0, +2, and +72 h) or MIP (+96 h). DAPI and antibody stainings for Sox2 and Bra. Scale bars, 150 μm. Dii: quantification of Chir treatment at 24 h (144 h quantification) or 48 h (120 h quantification). (n = 1,192 treated at 24 h, n = 807 treated at 48 h.) Diii: heatmaps: Z scored mean intensity of Sox2 and Bra.
(E) Bulk RNA sequencing (in triplicates) of gastruloids collected at 48, 72, and 96 h pulsed at indicated time points. Heatmap: Z scored expression levels of top 5 differentially expressed genes for cell-type annotations obtained from scRNA-seq. NMPs, neuro-mesodermal progenitors; PSM, pre-somitic mesoderm; PS, primitive streak; Def. endoderm, definitive endoderm; Exit. Naive pluripotency, exiting naive pluripotency.
(F) Scheme showing size regulation.
(G) Heatmaps: Z scored mean intensities corresponding to (H). (n = 170 gastruloids [150 cells], n = 239 gastruloids [300 cells], n = 215 gastruloids [500 cells]).
(H) Representative images at indicated cell number and time points. Middle z plane of z stack: DAPI and antibody stainings for Sox2, Fn1, and Bra. Scale bars, 150 μm.
(I) Scheme showing gastruloids seeded from different cell lines.
(J) Boxplots: comparisons of Pearson correlations of marker intensities (Sox2, Bra, and Fn1) stained in different cell lines (SBR, B/S [BramCh/Sox2Ve], E14, and Mesp1-GFP) at indicated time points. n = minimum 80 gastruloids per cell line and time point.
(K) Images corresponding to (J) of gastruloids from different cell lines (SBR, B/S [BramCh/Sox2Ve], E14, and Mesp1-GFP) fixed at 72, 96, and 120 h and stained for Bra, Sox2, and DAPI. Middle z plane for 72 h time point and MIPs for the other time points. Scale bars, 150 μm.