Figure 2.
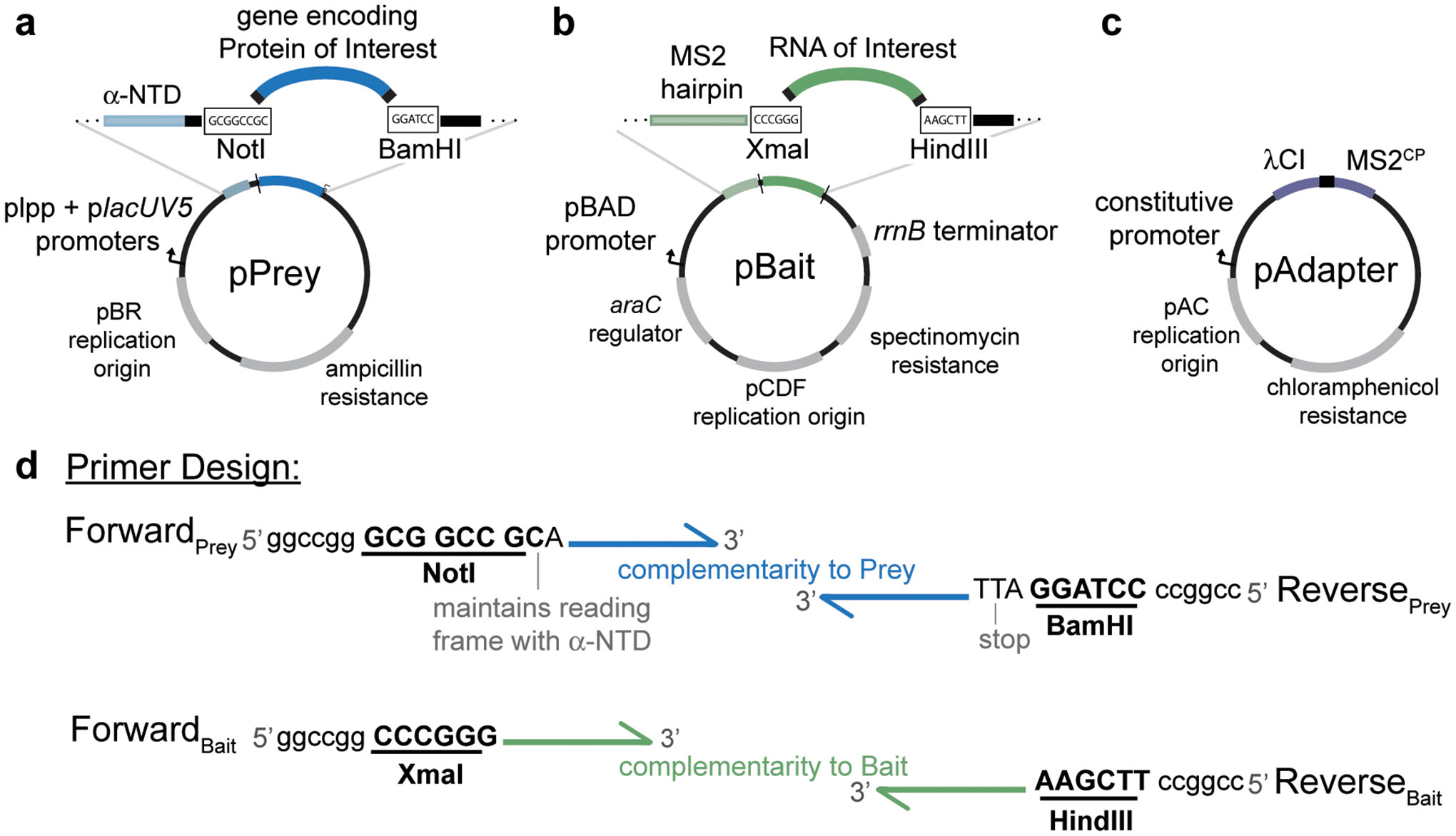
Plasmid maps of vectors used in B3H experiments highlighting key restriction sites used for cloning, promoters driving the expression of hybrid components and antibiotic resistance conferred by each plasmid. a, pPrey encodes the protein of interest tethered to α-NTD. Restriction enzymes NotI and BamHI downstream of α-NTD can be used for cloning different prey proteins into the plasmid. b, pBait encodes the hybrid-RNA containing an MS2hp and the RNA of interest. Restriction enzymes XmaI and HindIII downstream of MS2hp can be used for cloning different RNA baits into the plasmid. c, pAdapter encodes the DNA-RNA adapter fusion protein λCI-MS2CP. See Table 1 for additional plasmid details. d, Schematic representation of design of forward and reverse primers to clone a protein of interest into pPrey (top) and an RNA of interest into pBait (bottom). Restriction sites are bolded and underlined, and example “landing pads” to assist with restriction digestion of PCR products are shown at the 5’ ends of primers; these sequences provide additional dsDNA downstream of the restriction site, but the exact sequence of these is not critical. Note that an extra nucleotide following the NotI site is essential to maintain the reading frame of the α-NTD with the prey protein of interest. If you are cloning a domain of a protein that does not contain its own stop codon, one should be added upstream of the BamHI restriction site of the reverse primer.
