Abstract
Mitochondria play an important role in both normal heart function and disease etiology. We report analysis of common genetic variations contributing to mitochondrial and heart functions using an integrative proteomics approach in a panel of inbred mouse strains called the Hybrid Mouse Diversity Panel (HMDP). We performed a whole heart proteome study in the HMDP (72 strains, n=2-3 mice) and retrieved 848 mitochondrial proteins (quantified in ≥50 strains). High-resolution association mapping on their relative abundance levels revealed three trans-acting genetic loci on chromosomes (chr) 7, 13 and 17 that regulate distinct classes of mitochondrial proteins as well as cardiac hypertrophy. DAVID enrichment analyses of genes regulated by each of the loci revealed that the chr13 locus was highly enriched for complex-I proteins (24 proteins, P=2.2E-61), the chr17 locus for mitochondrial ribonucleoprotein complex (17 proteins, P=3.1E-25) and the chr7 locus for ubiquinone biosynthesis (3 proteins, P=6.9E-05). Follow-up high resolution regional mapping identified NDUFS4, LRPPRC and COQ7 as the candidate genes for chr13, chr17 and chr7 loci, respectively, and both experimental and statistical analyses supported their causal roles. Furthermore, a large cohort of Diversity Outbred mice was used to corroborate Lrpprc gene as a driver of mitochondrial DNA (mtDNA)-encoded gene regulation, and to show that the chr17 locus is specific to heart. Variations in all three loci were associated with heart mass in at least one of two independent heart stress models, namely, isoproterenol-induced heart failure and diet-induced obesity. These findings suggest that common variations in certain mitochondrial proteins can act in trans to influence tissue-specific mitochondrial functions and contribute to heart hypertrophy, elucidating mechanisms that may underlie genetic susceptibility to heart failure in human populations.
Research organism: Mouse
Introduction
Mitochondrial functions play a major role in the pathophysiology of several metabolic syndrome traits including obesity, insulin resistance and fatty liver disease (Yin et al., 2014; Chouchani et al., 2014; Begriche et al., 2006; Sanyal et al., 2001; Kim et al., 2008). There is now substantial evidence showing that genetic variation in mitochondrial functions contribute importantly to ‘complex’ diseases such as cardiovascular diseases (CVD) (Wallace, 2018). A scientific statement from the American Heart Association summarizes the central role of mitochondria in heart disease (Murphy et al., 2016). Mitochondrial bioenergetics and other functions impinge on virtually all aspects of the cell, including energy, cellular redox, apoptosis, and substrates for epigenetic modifications. Mitochondrial dysfunction also contributes to the development of heart failure (Brown et al., 2017; Zhou and Tian, 2018). The primary putative mechanism linking mitochondrial dysfunction to heart failure is decreased oxidative respiration leading to contractile failure. However, many other mechanisms have been postulated in recent years to implicate mitochondrial dysfunction in heart failure. They include excessive oxidative stress leading to inflammation, cell damage, and cell death; disturbed calcium homeostasis that triggers the opening of the mitochondrial permeability transition pore (mPTP), leading to loss of membrane potential, and eventual cell death. Therefore, mitochondria are an attractive target for heart failure therapy (Brown et al., 2017). Despite evidence showing that genetic variation in mitochondrial proteins is linked to disease, most studies tend to overlook the role of genetic variation in exploring the link between mitochondrial function and relevant phenotypes. To this end, we employ a ‘systems genetics’ approach to address this issue.
Our laboratory uses a combination of genetics, molecular biology, and informatics to investigate pathways underlying common cardiovascular and metabolic disorders. We exploit natural genetic variation among inbred strains of mice (and among human populations where possible) to identify novel targets and formulate hypotheses, taking advantage of unbiased global multi-omics technologies, such as transcriptomics, metabolomics, and proteomics, to help decipher causal mechanisms that drive complex traits. This ‘systems genetics’ approach integrates natural genetic variation with omics-level data (such as global protein abundance levels) to examine complex interactions that are difficult to address directly in humans. It shares with systems biology a holistic, global perspective (Civelek and Lusis, 2014; Seldin et al., 2019). Typically, a genetic reference population is examined for relevant clinical traits as well as global molecular traits, such as proteomics, and the data are integrated using correlation structure, genetic mapping, and mathematical modeling (Civelek and Lusis, 2014; Seldin et al., 2019).
Our systems genetics approach utilizes a well-characterized genetic reference population of 100 inbred mice strains, termed the Hybrid Mouse Diversity Panel (HMDP) (Lusis et al., 2016). The design of the HMDP resource, consisting of a panel of permanent inbred strains of mice that can be examined for many phenotypes, has proved invaluable for studying metabolic syndrome traits. Major advantages of this approach are that the mapping resolution for complex traits is superior to traditional genetic crosses and the use of inbred strains affords replication of biological measures. It also facilitates studies of gene-by-environment and gene-by-sex interactions, which are difficult to address in human populations. Using this resource, we have characterized several cardiometabolic traits in both sexes over the last 10 years (Parks et al., 2013; Parks et al., 2015; Rau et al., 2015; Org et al., 2016; Wang et al., 2016; Seldin et al., 2017; Rau et al., 2017; Norheim et al., 2017; Norheim et al., 2018; Hui et al., 2018; Seldin et al., 2018; Chella Krishnan et al., 2018; Chella Krishnan et al., 2019; Norheim et al., 2019; Chella Krishnan et al., 2021).
In the present study, we have explored the genetic regulation of mitochondrial pathways and their contribution to heart function in the HMDP. Using an integrative proteomics approach, we now report the identification of three independent genetic loci that control distinct classes of mitochondrial proteins as well as heart hypertrophy. Each locus contains proteins previously shown to affect heart pathophysiology but by unknown mechanisms. Our results show that genetic diversity in the mitochondrial proteome plays a central role in heart pathophysiology.
Results
Genetic architecture of the heart mitochondrial proteome revealed three trans-regulatory hotspots
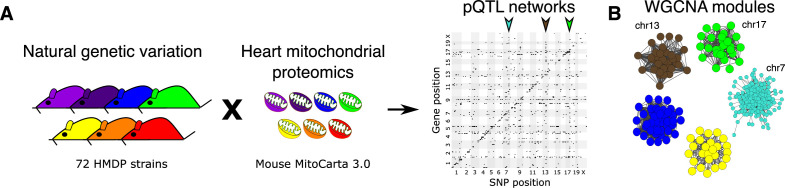
To investigate the effects of genetics on the heart proteome, we first performed a whole heart proteomic analysis in the HMDP (72 strains, n=2–3 mice, female sex; listed in Supplementary file 1a) and surveyed mitochondrial localization using MitoCarta3.0 (Rath et al., 2021). We retrieved abundance data for 848 of these proteins (quantified in ≥50 strains) and performed high-resolution association mapping on their respective abundance levels to the HMDP genotypes. Genetic variants associated to the protein abundance of nearby genes (<1 Mb) were referred to as cis- protein quantitative trait loci (pQTLs) and the remainder were referred to as trans-pQTLs. When a trans-pQTL locus is associated to multiple proteins, we defined it as a trans-pQTL hotspot. Using these criteria, we identified three hotspots, located on chromosome (chr) 7, chr13 and chr17, respectively (Figure 1A).
Figure 1. Genetic architecture of heart mitochondrial proteome.
(A) High-resolution association mapping of 848 heart mitochondrial proteins from 72 HMDP strains to identify pQTL networks. Associations between protein abundance levels and genetic variants located within 1 Mb of the respective gene location were considered as cis-pQTLs (p<1E-05) shown along the diagonal axis and the rest were considered as trans-pQTLs (p<1E-06). Three trans-pQTL hotspots are indicated by arrows. (B) Five WGCNA modules and the respective trans-pQTL hotspots are shown. HMDP, hybrid mouse diversity panel; pQTL, protein quantitative trait locus; WGCNA, weighted gene co-expression network analysis.
To identify the aspects of mitochondrial metabolism regulated by these loci, we constructed co-expression protein networks using weighted gene co-expression network analysis (WGCNA; Langfelder and Horvath, 2008) and identified five modules (Figure 1B and Supplementary file 1b). Eigengenes, representing the first principal component of two of these modules (Brown and Green), mapped to the same regions as the chr13 and chr17 loci, respectively. DAVID enrichment analyses (Huang et al., 2009) revealed that the chr13 locus (26 proteins; Figure 2A and Supplementary file 1c) that overlapped with Brown module (67 proteins, 96% overlap) was highly enriched for mitochondrial complex-I proteins (24 proteins, p=2.2E-61), and the chr17 locus (22 proteins; Figure 3A and Supplementary file 1d) that overlapped with Green module (38 proteins, 73% overlap) was highly enriched for mitochondrial ribonucleoprotein complex proteins (17 proteins, p=3.1E-25). The hotspot proteins in the chr7 locus (26 proteins; Figure 4A and Supplementary file 1e) were found primarily in the Turquoise module (402 proteins, 81% overlap) and was highly enriched for ubiquinone biosynthesis (3 proteins, p=6.9E-05).
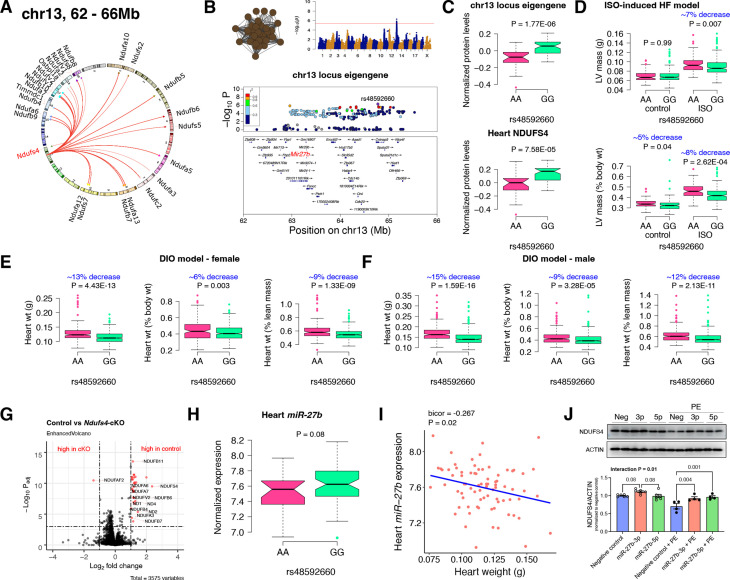
Figure 2. Chr13 locus controls mitochondrial complex-I via miR-27b/NDUFS4 axis.
(A) Circos plot showing chr13 hotspot. Each line signifies a significant association between the genetic variants and the respective protein levels with candidate genes being highlighted. (B) Manhattan and regional plots of chr13 locus (brown module) eigengene, respectively. Red line signifies genome-wide significance threshold (p<4.1E-06). The peak SNPs and the candidate genes are highlighted. Genotype distribution plots of (C) protein levels and eigengenes, and cardiac phenotypes from (D) ISO-induced HF model and (E) female and (F) male DIO model at peak SNP (rs48592660) associated with chr13 locus. (G) Volcano plot showing differentially expressed protein levels between control and heart-specific Ndufs4-cKO mice (n=5 mice/group). Significantly different proteins (significance cutoff: abs[log2FC]>1 and Padj <0.001) are highlighted in red. (H) Genotype distribution plots of heart miR-27b expression at peak SNP (rs48592660) associated with chr13 locus. (I) Gene-by-trait correlation plot between heart weight phenotype and heart miR-27b expression. (J) Immunoblot analyses of NDUFS4 protein levels in NRVMs transfected with mature miR-27b in the presence or absence of PE treatment. Data are presented as (C–F and H) boxplots showing median and interquartile range with outliers shown as circles (n=68–72 strains for protein levels; n=92–95 strains for ISO-model; n=92–100 strains for DIO-model; n=78 strains for miRNA levels) or (J) mean ± SEM (n=4–6 per group). p Values were calculated using (A and B) FaST-LMM that uses Likelihood-Ratio test; (C–F and H) Unpaired two-tailed Student’s t test; (I) BicorAndPvalue function of the WGCNA R-package that uses Unpaired two-tailed Student’s t test; (J) two-factor ANOVA corrected by post-hoc “Holm-Sidak’s” multiple comparisons test. ISO, isoproterenol; HF, heart failure; DIO, diet-induced obesity; NRVMs, neonatal rat ventricular myocytes; PE, phenylephrine.
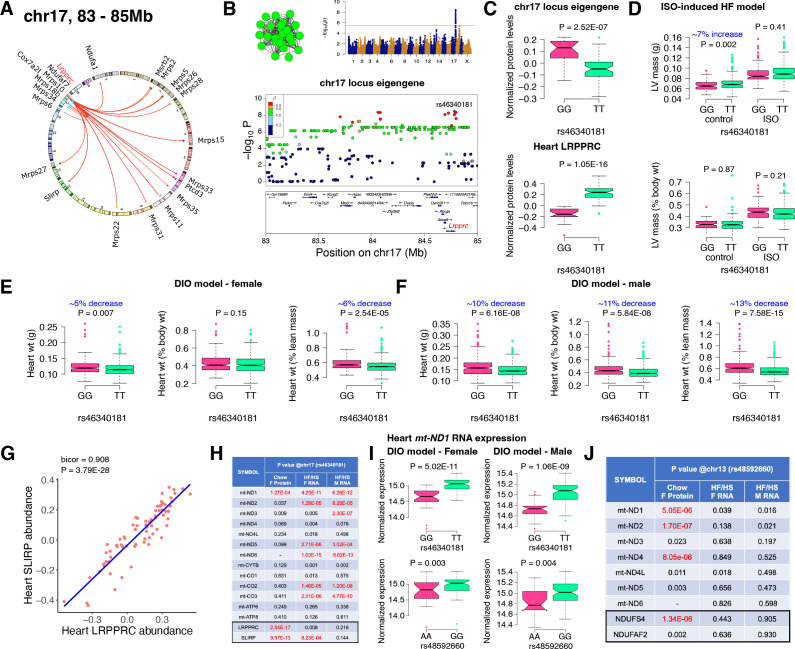
Figure 3. Chr17 locus controls mitoribosomes via LRPPRC/SLIRP.
(A) Circos plot showing chr17 hotspot. Each line signifies a significant association between the genetic variants and the respective protein levels with candidate genes being highlighted. (B) Manhattan and regional plots of chr17 locus (green module) eigengene, respectively. Red line signifies genome-wide significance threshold (P<4.1E-06). The peak SNPs and the candidate genes are highlighted. Genotype distribution plots of (C) protein levels and eigengenes, and cardiac phenotypes from (D) ISO-induced HF model and (E) female and (F) male DIO model at peak SNP (rs46340181) associated with chr17 locus. (G) Protein-by-protein correlation plot between heart LRPPRC and SLIRP abundance levels. (H) Association p values between mtDNA-encoded mRNA expression or protein abundance levels and peak SNP (rs46340181) associated with chr17 locus in both sexes of HMDP. (I) Genotype distribution plots of heart mt-ND1 mRNA expression from female and male DIO model at peak SNPs associated with chr17 (rs46340181) or chr13 (rs48592660) loci, respectively. (J) Association p values between mtDNA-encoded complex-I related mRNA expression or protein abundance levels and peak SNP (rs48592660) associated with chr13 locus in both sexes of HMDP. Data are presented as (C–F and I) boxplots showing median and interquartile range with outliers shown as circles (n=68–72 strains for protein levels; n=92–95 strains for ISO-model; n=92–100 strains for DIO-model). p Values were calculated using (A, B, H and J) FaST-LMM that uses Likelihood-Ratio test; (C–F and I) Unpaired two-tailed Student’s t test; (G) BicorAndPvalue function of the WGCNA R-package that uses Unpaired two-tailed Student’s t test.
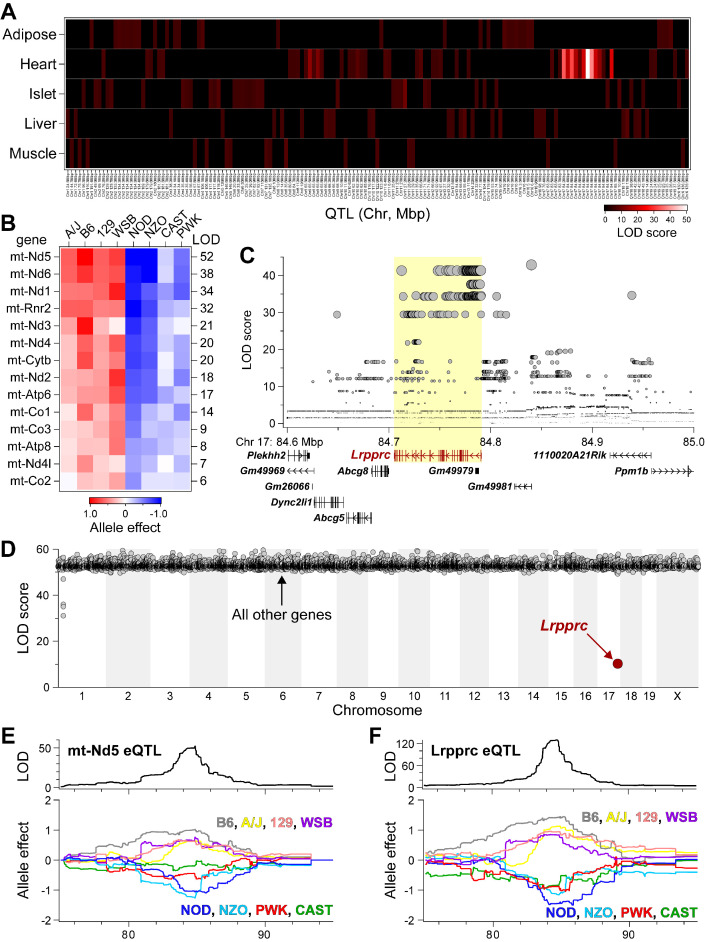
Figure 4. Lrpprc regulates an mt-encoded eQTL hotspot specifically in heart of DO mice.
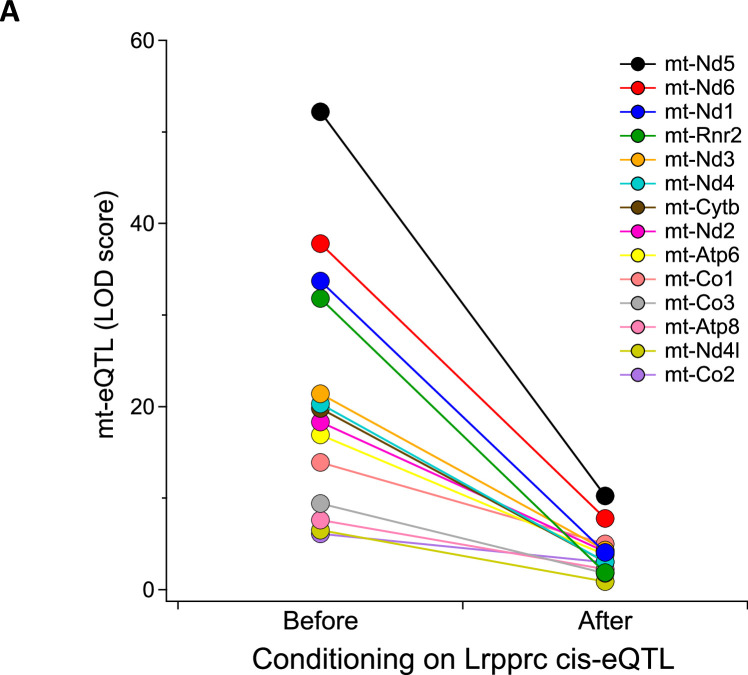
(A) Heatmap illustrates all eQTL (LOD >6) for mt-encoded transcripts that were identified in Adipose (Wang et al., 2012), Heart (Ruzzenente et al., 2012), Islet (Gu et al., 2016), Liver (Rath et al., 2021), and Muscle (Wang et al., 2016) from Diversity Outbred mice maintained on a Western-style diet. Mt-eQTL are arranged along x-axis according to genomic position from chr1 to chrX; z-axis depicts LOD score, highlighting a hotspot on chr17 at ~85 Mbp. (B) Allele effect patterns for mt-eQTL mapping to chr17 hotspot in heart. Red illustrates alleles associated with increased expression; blue, decreased expression. LOD scores are shown along right margin. (C) SNP association profile for mt-Nd5 eQTL in heart. Lrpprc contains SNPs with strongest association, yellow highlighted region. (D) Mediation of mt-Nd5 eQTL against all transcripts in heart. The LOD score for mt-Nd5 is significantly reduced when conditioned on a Lrpprc cis-eQTL, consistent with genetic regulation of Lrpprc being required for the regulation of mt-Nd5. The mt-Nd5 eQTL (E) and Lrpprc cis-eQTL (F) in heart demonstrate matched and concordant allele effect patterns, suggesting Lrpprc is a positive driver of mt-Nd5.
Figure 4—figure supplement 1. Mediation analysis identifies Lrpprc as a driver for the heart-specific mt- eQTL hotspot on chr17.
Chr13 locus controls mitochondrial complex-I
First, we analyzed the chr13 locus (26 proteins) that was highly enriched for mitochondrial complex-I (Figure 2A). Mapping the eigengene of the chr13 trans-regulated proteins identified the peak SNP (rs48592660). NDUFS4, a protein critical for complex-I assembly and loss of which leads to cardiac hypertrophy (Chouchani et al., 2014) mapped near the locus but outside the region of linkage disequilibrium. However, within the locus was the microRNA(miR)–23b/27b/24–1 cluster, among which miR-27b, a conserved regulator of NDUFS4, was identified via nine miRNA target prediction algorithms and one dataset of experimentally validated miRNA targets (Figure 2B and Supplementary file 1f). Cardiac overexpression of miR-27b has previously been shown to promote cardiac hypertrophy (Wang et al., 2012) but attenuate angiotensin II-induced atrial fibrosis (Wang et al., 2018). We therefore hypothesized that the chr13 locus regulated complex-I proteins by influencing miR-27b and thus NDUFS4 protein levels. Indeed, we observed higher levels of both the chr13 locus eigengene and NDUFS4 with the GG allele of the peak locus SNP (Figure 2C). To identify the functional relevance of this peak SNP, we analyzed its association in two independent heart stress models, namely, isoproterenol (ISO)-induced heart failure (Wang et al., 2016) and diet-induced obesity (DIO) (Parks et al., 2013) models. We observed significantly lower left ventricular mass under ISO stress (Figure 2D) and lower heart weight under DIO stress in both sexes (Figure 2E) of strains harboring the GG allele, thus confirming the directionality of genetic impacts on NDUFS4 protein levels and hypertrophic response.
NDUFS4 heart-specific knockout mice had reduced mitochondrial complex-I proteins
NDUFS4 is an 18 kDa accessory subunit that is essential for the mitochondrial complex-I assembly (Zhu et al., 2016; Stroud et al., 2016; Gu et al., 2016; Kahlhöfer et al., 2017; Scacco et al., 2003). Loss-of-function mutations in NDUFS4 leads to complex-I deficiency causing a neuromuscular disease, Leigh syndrome (Leshinsky-Silver et al., 2009) and is also involved in cardiomyopathies (Chouchani et al., 2014; Karamanlidis et al., 2013; Zhang et al., 2018). Taken together, we wanted to experimentally test our hypothesis that NDUFS4 protein independently controls the complex-I protein abundance levels in heart. For this, we performed whole heart proteomic analyses in both control and heart-specific Ndufs4-cKO mice (n=5 mice/group). Among the abundance data for 3575 proteins, we observed 31 proteins to be significantly different between the control and cKO groups (significance cutoff: abs[log2FC]>1 and Padj <0.001; listed in Supplementary file 1g). Strikingly, 21 of these proteins were found in the chr13 locus and these were highly enriched for complex-I proteins (30 proteins, p=3.8E-80). Notably, only one protein, NDUFAF2, was up regulated in Ndufs4-cKO mice (Figure 2G). This is intriguing because NDUFAF2 has been reported to stabilize complex-I in the absence of NDUFS4 and to sustain complex-I activity (Adjobo-Hermans et al., 2020; Leong et al., 2012) indicating this may be a compensatory mechanism.
MiR-27b controls NDUFS4 protein levels and heart weights
As an independent corroboration and to understand the consequence of the chr13 locus on miR-27b expression, we independently sequenced miRNAs from DIO-stressed female HMDP strains (n=78 strains). We found that strains harboring the GG allele had higher miR-27b expression (Figure 2H), and there was a significant inverse correlation between heart weights and miR-27b expression in these mice (Figure 2I). Based on these observations, we hypothesize that miR-27b increases NDUFS4 protein levels thereby reducing heart weights. To validate this observation, we transfected neonatal rat ventricular myocytes (NRVMs) with mature miR-27b in the presence or absence of phenylephrine (PE) treatment. The precursor miR-27b has two mature arms: miR-27b-5p and miR-27b-3p that originates from the 5’ or 3’ strand of the precursor miR-27b, respectively and are almost complementary to each other. The stability and functionality of each arm depends on the tissue/cell type and according to miRBase Sequence database (Kozomara and Griffiths-Jones, 2011) (database of published miRNA sequences and annotation), miR-27b-3p (98.8%) is more abundant than miR-27b-5p (1.2%) and therefore is assumed to be the stable and functional version. Immunoblot analyses revealed that NDUFS4 protein levels were reduced with PE treatment in the control cells but miR-27b-3p consistently increased NDUFS4 protein levels in both control and PE-treated conditions while miR-27b-5p increased NDUFS4 protein levels only in the PE-treated conditions (Figure 2J). Taken together, we conclude that the chr13 locus affects mitochondrial complex-I proteins through the miR-27b/NDUFS4 axis, thereby controlling heart weights.
Chr17 locus controls mitoribosomes
Next, we analyzed the chr17 locus (22 proteins) that was highly enriched for mitochondrial ribosomal proteins (Figure 3A). We mapped the eigengene of the significantly associated mitochondrial proteins to identify the peak SNP (rs46340181). We identified LRPPRC as a candidate as it was the only protein controlled in cis by the peak SNP (Figure 3B). Importantly, LRPPRC together with SLIRP controls mitochondrial mRNA stability, enabling polyadenylation and translation (Siira et al., 2017; Chujo et al., 2012; Ruzzenente et al., 2012). Loss-of-function mutations in LRPPRC cause a congenital mitochondrial disease called Leigh syndrome, French-Canadian type that is often characterized by mitochondrial complex IV deficiency and impaired mitochondrial respiration and in some cases, neonatal cardiomyopathy and congenital cardiac abnormalities have been reported (Mootha et al., 2003; Oláhová et al., 2015). It is noteworthy that SLIRP is also under the control of the chr17 locus (Figure 3A). Further, our phenotypic associations revealed that the eigengene and LRPPRC were inversely associated with the TT allele (Figure 3C). This was functionally translated into lower heart weight in strains harboring the TT allele in both sexes under DIO stress only (Figure 3D–F). We also observed that abundance levels of both LRPPRC and SLIRP proteins were strongly correlated with each other (Figure 3G) and controlled by the chr17 locus (Figure 3A), thus demonstrating that they are co-regulated.
LRPPRC/SLIRP protein complex controls mitochondrial transcript levels
Based on our current observations and published data, we hypothesized that high LRPPRC/SLIRP protein complex stabilizes mitochondrial transcripts, thus reducing the need to upregulate mitochondrial translation. To test this, we independently sequenced the heart transcripts from our HMDP mice that underwent DIO stress. We observed that the chr17 locus peak SNP (rs46340181), which strongly controls both LRPPRC and SLIRP proteins, is only associated with the mitochondrial mRNA expression and not their respective protein levels (Figure 3H). Moreover, this phenomenon was observed in both sexes, explaining the lack of sex bias in phenotypic associations (Figure 3E and F). In contrast, the chr13 locus that controlled complex-I proteins did not show strong associations with transcript levels in either sex (Figure 3I and J). As a specific example, Figure 3I shows that the mRNA expression of mt-ND1 was higher in strains harboring the TT allele in both sexes under DIO stress, illustrating that upregulated LRPPRC/SLIRP is stabilizing the mitochondrial transcript (Figure 3C), resulting in reduced heart weights (Figure 3D–F).
Corroboration with Diversity Outbred mice also identifies a heart-specific chr17 mt-eQTL hotspot controlled by LRPPRC
As independent corroboration, we utilized the Diversity Outbred (DO) mice (Svenson et al., 2012) to further explore our chr17 locus. To extend our observations about the genetics of mitochondrially encoded (mt-encoded) gene regulation, we asked if mt-encoded genes are subjected to genetic regulation in multiple tissues from a large cohort of DO mice. We used RNA-sequencing to survey gene expression that included transcripts encoded by the mitochondrial genome, in five tissues (adipose, heart, islet, liver and skeletal muscle) from ~500 DO mice that were maintained on a Western-style diet high in fat and sucrose. All DO mice were genotyped at >140 K SNPs with the GigaMUGA microarray (Morgan et al., 2015), enabling eQTL analysis of the mt-encoded transcripts. Among the five tissues surveyed, we identified 157 mt-eQTL (LOD >6) for 15 mt-encoded genes, including all 13 protein-coding genes, and the two genes that encode for ribosomal RNA proteins (Supplementary file 1h). Given that the mitochondrial and nuclear genomes are distinct, mt-encoded eQTL at nuclear loci reflect trans-acting mechanisms that bridge the two genomes (Ali et al., 2019).
While all tissues yielded multiple mt-eQTL, heart demonstrated a striking hotspot where 14 mt-encoded genes uniquely mapped to chr17 at ~85 Mbp with LOD scores ranging from 52 (mt-Nd5) to 6 (mt-Co2) (Figure 4A; listed in Supplementary file 1h). The founder allele-effect signatures for the 14 mt-eQTL at the hotspot were the same and partitioned the eight haplotypes into ‘high’ (A/J, B6, 129, WSB) versus ‘low’ (NOD, NZO, CAST, PWK) subgroups (Figure 4B), suggesting a common causal variant behind the co-mapping of the mt-eQTL. The SNP association profile for the mt-Nd5 eQTL identified a block of SNPs directly over the Lrpprc gene locus (Figure 4C), including one missense variant (rs33393440, Lys466Glu), which is present in the high allele subgroup (Supplementary file 1i). Taken together, these results suggest that SNPs within Lrpprc may be responsible for the heart-specific mt-eQTL hotspot.
We used mediation analysis to identify potential causal gene underlying physiological or molecular QTL as described (Keller et al., 2016; Keller et al., 2018; Keller et al., 2019). In mediation analysis of gene expression, trans-eQTL are conditioned on the expression of all other genes, including those at the locus to which the trans-eQTL map. If the genetic signal of the trans-eQTL decreases upon conditioning of a specific gene, that gene becomes a strong candidate as the driver of the trans-eQTL. We focused on the trans-eQTL for mt-Nd5 at the chr17 locus in heart, as this demonstrated the strongest genetic signal (Figure 4B). Mediation against Lrpprc resulted in a large drop in the LOD score for mt-Nd5 eQTL (Figure 4D). Similar results were observed for mediation of the other mt-eQTL to the heart hotspot (Figure 4—figure supplement 1). The allele effect patterns of the mt-Nd5 eQTL (Figure 4E) and that for the Lrpprc eQTL (Figure 4F) demonstrate the same, and concordant high and low haplotype subgroups. Taken together, our results in DO mice are consistent with that in the HMDP mouse cohort, and strongly suggest that LRPPRC functions as a positive driver of the mt-eQTL hotspot in heart.
Chr7 locus affects CoQ metabolism
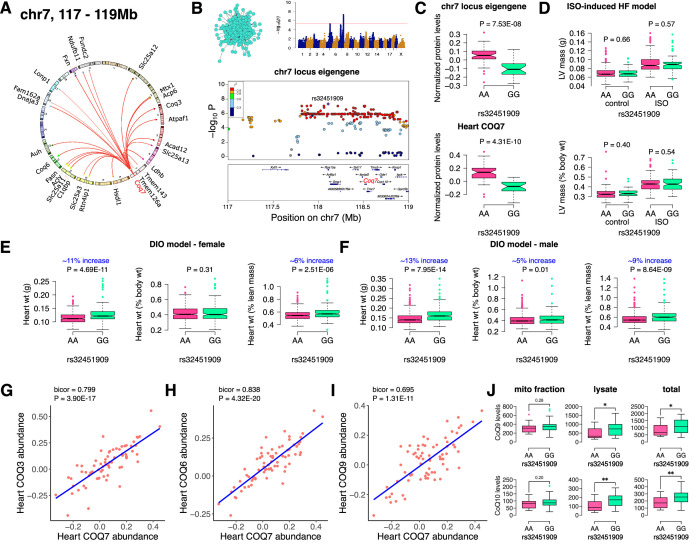
Finally, we analyzed the chr7 locus (26 proteins), which unlike the chr13 or chr17 loci, had no major representation of a single mitochondrial protein complex but was moderately enriched for ubiquinone biosynthesis (Figure 5A). Mapping the eigengene of the significantly associated mitochondrial proteins identified the peak SNP (rs32451909). We identified COQ7 as a strong candidate as it was the only protein exhibiting a cis-regulation at the locus in the region of linkage disequilibrium (Figure 5B). COQ7 catalyzes a critical step in the biosynthesis of coenzyme Q (CoQ). Among several functions, CoQ participates in electron transport facilitating ATP synthesis. CoQ also has a clear role in heart failure (Sharma et al., 2016). Phenotypically, we observed lower levels of both the eigengene and COQ7 with the GG allele (Figure 5C). This was functionally translated into higher heart weight in strains containing GG allele in both sexes under DIO stress only (Figure 5D–F). We also observed that abundance levels of other COQ proteins were strongly correlated with COQ7 protein, thus demonstrating that they are co-regulated (Figure 5G–I). At least two of these proteins, COQ3 and COQ6, are associated with the chr7 locus (Figure 5A). When we measured the CoQ levels in both the mitochondrial fractions and total heart lysates from DIO-stressed female HMDP strains (n=15 strains), we observed a significant upregulation in the levels of both CoQ9 and CoQ10 in the lysates from HMDP mice harboring the GG allele (Figure 5J). Taken together, we conclude that the chr7 locus controls CoQ metabolism via regulation of the COQ7 protein.
Figure 5. Chr7 locus affects CoQ metabolism via COQ7.
(A) Circos plot showing chr7 hotspot. Each line signifies a significant association between the genetic variants and the respective protein levels with candidate genes being highlighted. (B) Manhattan and regional plots of chr7 locus (turquoise module) eigengene, respectively. Red line signifies genome-wide significance threshold (p<4.1E-06). The peak SNPs and the candidate genes are highlighted. Genotype distribution plots of (C) protein levels and eigengenes, and cardiac phenotypes from (D) ISO-induced HF model and (E) female and (F) male DIO model at peak SNP (rs32451909) associated with chr7 locus. Protein-by-protein correlation plots between heart COQ7 and (G) COQ3, (H) COQ6 and (I) COQ9 abundance levels. (J) Genotype distribution plots of CoQ9 or CoQ10 levels in both the mitochondrial fractions and total heart lysates from female DIO model at peak SNPs associated with chr7 (rs32451909) locus. Data are presented as (C–F and J) boxplots showing median and interquartile range with outliers shown as circles (n=68–72 strains for protein levels; n=92–95 strains for ISO-model; n=92–100 strains for DIO-model; n=15 strains for CoQ9 and CoQ10 levels). p Values were calculated using (A and B) FaST-LMM that uses Likelihood-Ratio test; (C–F and J) Unpaired two-tailed Student’s t test; (G–I) BicorAndPvalue function of the WGCNA R-package that uses Unpaired two-tailed Student’s t test.
Discussion
We have previously used systems genetics analyses in HMDP and discovered a central role for adipose mitochondrial function in the sex differences observed in cardiometabolic traits, including obesity, insulin resistance and plasma lipids (Norheim et al., 2019; Chella Krishnan et al., 2021), and liver mitochondrial function in non-alcoholic fatty liver disease (Chella Krishnan et al., 2018). In the present study, an integrative proteomics approach was utilized in HMDP to investigate the effects of genetic regulation of mitochondrial pathways on heart function. Several conclusions have emerged. First, we identified three distinct and independent trans-regulating genetic loci, on chr13, chr17 and chr7, respectively. Second, our enrichment analyses suggested mechanisms perturbed by each of these loci. Thus, the chr13 locus was enriched for mitochondrial complex I, chr17 for mitoribosomes, and chr7 was enriched for ubiquinone biosynthesis. Third, our regional mapping identified NDUFS4, LRPPRC and COQ7 as candidate genes for chr13, chr17, and chr7 loci, respectively. Finally, we performed both experimental and statistical analyses to support their causal roles. Each of these points is discussed in detail below.
Trans-acting human pQTLs, mostly using blood/plasma proteome (Yao et al., 2018; Ruffieux et al., 2020; Zhong et al., 2020; Yang et al., 2021; Folkersen et al., 2020; He et al., 2020), and trans-acting human eQTLs (Aguet, 2017; Brynedal et al., 2017; Yao et al., 2017; Small et al., 2018; GTEx Consortium, 2020), have been reported, but exploring trans-regulatory landscapes in human studies is limited by sample size, tissue accessibility, and environmental factors. In the current study, mapping genetic loci controlling protein abundance levels in heart tissues from an inbred mouse population revealed that ~9% of available nuclear-encoded mitochondrial proteins (74/848) are significantly controlled (p<1E-6) by three genetic loci, on chr13 (26 proteins), chr17 (22 proteins) and chr7 (26 proteins), respectively. Each of these three trans regulating hotspots were specific for the proteome, as they were not identified in our heart transcriptome. Interestingly, two of these hotspots independently controlled mitochondrial complex I, one at the mRNA (chr17 via LRPPRC/SLIRP) and the other at the protein level (chr13 via miR-27b/NDUFS4). Finally, the chr7 locus was found to control ubiquinone metabolism via COQ7 protein.
Mitochondrial complex I is the first and largest (~1 MDa) protein complex of the mitochondrial electron transport chain comprised of 45 subunits, seven encoded in the mtDNA (Zhu et al., 2016; Fiedorczuk et al., 2016; Hirst, 2013). NDUFS4 is an 18 kDa accessory subunit that assembles the catalytic N-module (where NADH oxidation occurs) with the rest of the complex I. Mutations leading to the loss of NDUFS4 result in complex I deficiency (Zhu et al., 2016; Stroud et al., 2016; Gu et al., 2016; Kahlhöfer et al., 2017; Scacco et al., 2003) and are involved in cardiomyopathies, both in protective (ischemia/reperfusion injury) and detrimental roles (pressure overload) (Chouchani et al., 2014; Karamanlidis et al., 2013; Zhang et al., 2018). It was also reported that loss of NDUFS4 causes reduced complex I mediated ROS generation (ischemia/reperfusion injury) and increased protein acetylation (pressure overload), leading to their respective pathologies. Here, we demonstrate that heart-specific NDUFS4 KO mice have higher levels of NDUFAF2, which is reported to partly stabilize complex I in the absence of NDUFS4, although the resultant complex I is unstable and has reduced activity (Adjobo-Hermans et al., 2020). Taken together, we report that natural variation in NDUFS4 protein levels affect other complex I proteins resulting in reduced complex I activity and cardiac hypertrophy. On the other hand, the role of miR-27b on heart function is unclear. Cardiac-specific overexpression of miR-27b has been reported to increase pressure overload-induced cardiac hypertrophy via PPARG (Wang et al., 2012) but attenuated angiotensin II-induced atrial fibrosis via ALK5 (Wang et al., 2018). Also, whole body knockout of miR-27b was found to attenuate pressure overload-induced cardiac hypertrophy via FGF1 (Li et al., 2022), but this could be due to extracardiac effects mediated by miR-27b-3p such as adipocyte browning (Yu et al., 2018; Sun and Trajkovski, 2014; Kong et al., 2015). Using 100 HMDP inbred strains of mice, here we found that cardiac miR-27b expression was inversely correlated with heart weights and that mature miR-27b-3p appears to have a protective role in reducing cardiac hypertrophy via increasing NDUFS4 protein levels.
Mitochondria are unique organelles having their own genome that encodes for 13 protein coding genes, which are essential subunits of complexes I, III, IV, and V. Nuclear-encoded LRPPRC protein working together with the SLIRP protein helps to enable mitochondrial mRNA translation via promoting its polyadenylation and stability (Siira et al., 2017; Chujo et al., 2012; Ruzzenente et al., 2012). Loss-of-function mutations in LRPPRC causes Leigh syndrome, French-Canadian type that are associated with complex IV deficiency, impaired mitochondrial respiration and in some cases, neonatal cardiomyopathy, and congenital cardiac abnormalities (Mootha et al., 2003; Oláhová et al., 2015). Here, we show that natural genetic variations in a panel of inbred mouse strains at a chr17 locus (TT allele) result in higher protein levels of the LRPPRC/SLIRP complex as well as lower heart weights. This phenomenon is most likely mediated by increased expression levels of mitochondrial mRNA that coincide with reduced protein levels of mitoribosomes. We propose that since these mitochondrial mRNAs are protected by LRPPRC/SLIRP complex, fewer mitoribosomes are necessary for translation. Interestingly, although LRPPRC mutations are often associated with complex IV deficiency (Mootha et al., 2003; Oláhová et al., 2015), we observed that complex I mRNAs were also strongly regulated by the LRPPRC/SLIRP complex, in addition to complex IV mRNAs. Thus, further research on the bioenergetic consequences of LRPPRC/SLIRP loss-of-function mutations beyond complex IV deficiency are warranted.
Mitochondrial coenzyme Q, or ubiquinone or CoQ, is essential for mitochondrial electron transport chain function as it shuttles electrons from both complexes I and II to complex III. The enzyme COQ7 is responsible for catalyzing the penultimate step in CoQ biosynthesis. Here, we report that the chr7 locus (GG allele) is associated with reduced levels of COQ7 protein, which is in turn associated with increased heart weights. Interestingly, when we measured CoQ levels in these hearts, we found no differences in the mitochondrial fractions but an increase in both CoQ9 and CoQ10 levels in the lysates. Mice with a complete loss of Coq7 expression have embryonic lethality (Nakai et al., 2001) but survive for several months after a knockout is induced in the adult (Wang et al., 2015). Interestingly, Coq7 heterozygous knockout mice are long-lived but exhibit dysfunctional mitochondria (such as reduced respiration, reduced ATP levels and increased ROS generation) with no differences in CoQ levels despite significantly reduced COQ7 protein levels (Lapointe and Hekimi, 2008; Lapointe et al., 2009; Liu et al., 2005). Furthermore, these Coq7 heterozygous knockout mice were found to harbor a varying distribution of CoQ9 levels in their liver mitochondria with the outer membrane having increased CoQ9 while the inner membrane had reduced CoQ9 levels (Lapointe et al., 2012). It was postulated that higher CoQ9 levels in the liver mitochondrial outer membrane may be a protective response to increased ROS levels generated in Coq7 heterozygous knockout mice (Lapointe et al., 2012). Thus, reduced COQ7 protein levels do not necessarily associate with reduced CoQ levels but have heterogeneous sub-mitochondrial and possibly subcellular CoQ distributions. Taken together, we propose that increased heart weights in mice harboring chr7 locus (GG allele) is a result of increased ROS generation caused by reduced COQ7 protein levels. Also, the increased CoQ levels in the heart lysates of these mice might be a protective response against the oxidative stress generated by reduced COQ7 protein in these mice. Further research is warranted in understanding the purported role of COQ7 in submitochondrial and subcellular CoQ heterogeneous distribution focusing on heart pathophysiology.
In conclusion, our unbiased systems genetics analyses identified three loci regulating mitochondrial function in the heart. None of these loci were observed when transcript levels were examined, providing justification for proteomic rather than transcriptomic studies. All three loci are associated with heart mass in two independent heart stress models and/or negatively correlated with known hypertrophic markers (Supplementary file 1j). Nevertheless, the percent change in heart mass between the three loci were only of modest value. However, both heart failure and hypertrophy being complex traits are influenced by a large number of genetic factors, each exerting a small to large effect. The resulting phenotypes are therefore a sum of all impacts. So, we believe it is important to characterize modest as well as large effects. In summary, our results provide mechanistic information about the roles of previously studied genes namely, NDUFS4, LRPPRC, and COQ7, in heart failure.
Methods
Mice
All mice were purchased from The Jackson Laboratory and bred at UCLA according to approved institutional animal care and use committee (IACUC) protocols with daily monitoring. Both the ISO-induced HF (Rau et al., 2015; Wang et al., 2016; Rau et al., 2017) and DIO (Parks et al., 2013; Parks et al., 2015) models were previously described in detail. Briefly, for ISO-induced HF model, 8–10 weeks of age female mice were administered with isoproterenol (30 mg per kg body weight per day, Sigma) for 21 days using ALZET osmotic minipumps, which were surgically implanted intraperitoneally. For DIO model, mice were fed ad libitum a chow diet (Ralston Purina Company) until 8 weeks of age and then placed ad libitum on a high-fat/high-sucrose (HF/HS) diet (Research Diets-D12266B, New Brunswick, NJ) with 16.8% kcal protein, 51.4% kcal carbohydrate, 31.8% kcal fat for an additional 8 weeks. For heart proteomic analysis, 8–12 weeks of age female HMDP mice (72 strains, n=2–3 mice; listed in Supplementary file 1a) and 3–5 months old control and Ndufs4-cKO mice of both sex (n=5 each group) fed with chow diet ad libitum (Purina 5053, LabDiet) were used. All mice were maintained on a 14 hr light/10 hr dark cycle (light is on between 6 a.m. and 8 p.m.) at a temperature of 25 degrees and 30–70% humidity. On the day of the experiment, the mice were sacrificed after 4 hr fasting.
Studies in Diversity Outbred mice
All experiments that used Diversity Outbred (DO) mice were conducted in the Biochemistry Department at the University of Wisconsin-Madison and approved by the university’s Animal Care and Use Committee as described previously (Keller et al., 2018). Briefly, 4-week-old DO mice were purchased from the Jackson Labs (stock no. 009376) and placed on a Western-style diet high in fat and sucrose (17.3% protein, 34% carbohydrate, and 44.6% kcal fat) from Envigo Teklad (TD.08811). Mice were sacrificed at ~20 weeks of age, and tissues harvested and prepared for RNA-sequencing and expression quantitative trait loci (eQTL) analysis as previously detailed (Keller et al., 2018). The expression of the 13 protein-coding mt-genes (mt-Nd1, mt-Nd2, mt-Nd3, mt-Nd4, mt-Nd4l, mt-Nd5, mt-Nd6, mt-Cytb, mt-Co1, mt-Co2, mt-Co3, and mt-Atp6, mt-Atp8) and two mt-ribosomal RNAs (mt-Rnr1, mt-Rnr2) were detected in each of the five tissues surveyed (heart, islet, skeletal muscle, adipose, and liver). Expression values for these mt-encoded transcripts in all tissues, and Lrpprc in heart, are provided in Supplementary file 2.
Heart global proteomic analysis
Heart tissue from the HMDP, control and Ndufs4-cKO mice were lysed in 6 M guanidine HCL (Sigma; #G4505), 100 mM Tris pH 8.5 containing 10 mM tris(2-carboxyethyl)phosphine (Sigma; #75259) and 40 mM 2-chloroacetamide (Sigma; #22790) by tip-probe sonication. The lysate was heated at 95 °C for 5 min and centrifuged at 20,000 x g for 10 min at 4 °C. The supernatant was diluted 1:1 with water and precipitated overnight with five volumes of acetone at –20 °C. The lysate was centrifuged at 4000 x g for 5 min at 4 °C and the protein pellet was washed with 80% acetone. The lysate was centrifuged at 4000 x g for 5 min at 4 °C and the protein pellet was resuspended in Digestion Buffer (10% 2,2,2-Trifluoroethanol [Sigma; #96924] in 100 mM HEPEs pH 8.5). Protein was quantified with BCA (ThermoFisher Scientific) and normalized in Digestion Buffer to a final concentration of 2 µg/µl. Protein was digested with sequencing grade trypsin (Sigma; #T6567) and sequencing grade LysC (Wako; #129–02541) at a 1:50 enzyme:substrate ratio overnight at 37 °C with shaking at 2000 x rpm. Eight micrograms of peptide was directly labelled with 32 µg of 10-plex TMT (lot #QB211242) in 20 µl at a final concentration of 50% acetonitrile for 1.5 hr at room temperature. The reaction was de-acylated with a final concentration of 0.3% (w/v) hydroxylamine and quenched with a final concentration of 1% trifluoroacetic acid (TFA). Each 10-plex experiment contained nine different strains with a tenth reference label (131 isobaric label) made up of the same peptide digest from pooled mix of C57BL/6 J heart. Following labelling, the peptides from each of the 18 TMT 10-plex batches were pooled and purified directly by Styrene Divinylbenzene - Reversed-Phase Sulfonate (SDB-RPS) microcolumns, washed with 99% isopropanol containing 1% TFA and eluted with 80% acetonitrile containing 2% ammonium hydroxide followed by vacuum concentration. Peptides were resuspended in 2% acetonitrile containing 0.1% TFA and thirty micrograms of peptide was fractionated on an in-house fabricated 25 cm x 320 µm column packed with C18BEH particles (3 µm, Waters). Peptides were separated on a gradient of 0–30% acetonitrile containing 10 mM ammonium formate (pH 7.9) over 60 min at 6 µl/min using an Agilent 1260 HPLC and detection at 210 nm with a total of 48 fractions collected and concatenated down to 12 fractions.
Mass spectrometry and data processing
Peptide fractions from heart were resuspended in 2% acetonitrile containing 0.1% TFA and analyzed on a Dionex ultra-high pressure liquid chromatography system coupled to an Orbitrap Lumos mass spectrometer. Briefly, peptides were separated on 40 cm x 75 µm column containing 1.9 µm C18AQ Reprosil particles on a linear gradient of 2–30% acetonitrile over 2 h. Electrospray ionization was performed at 2.3 kV with 40% RF lens and positively charged peptides detected via a full scan MS (350–1550 m/z, 1e6 AGC, 60 K resolution, 50ms injection time) followed data-dependent MS/MS analysis performed with CID of 35% normalized collision energy (NCE) (rapid scan rate, 2e4 AGC, 50ms injection time, 10ms activation time, 0.7 m/z isolation) of the top 10 most abundant peptides. Synchronous-precursor selection with MS3 (SPS-MS3) analysis was enabled with HCD of 60 NCE (100–500 m/z, 50 K resolution, 1e5 AGC, 105ms injection time) (McAlister et al., 2014). Dynamic exclusion was enabled for 60 s. Data were processed with Proteome Discoverer v2.3 and searched against the Mouse UniProt database (November 2018) using SEQUEST (Eng et al., 1994). The precursor MS tolerance were set to 20 ppm and the MS/MS tolerance was set to 0.8 Da with a maximum of 2 miss-cleavage. The peptides were searched with oxidation of methionine set as variable modification, and TMT tags on peptide N-terminus / lysine and carbamidomethylation of cysteine set as a fixed modification. All data was searched as a single batch and the peptide spectral matches (PSMs) of each database search filtered to 1% FDR using a target/decoy approach with Percolator (Käll et al., 2007). The filtered PSMs from each database search were grouped and q-values generated at the peptide level with the Qvality algorithm (Käll et al., 2009). Finally, the grouped peptide data was further filtered to 1% protein FDR using Protein Validator. Quantification was performed with the reporter ion quantification node for TMT quantification based on MS3 scans in Proteome Discoverer. TMT precision was set to 20 ppm and corrected for isotopic impurities. Only spectra with <50% co-isolation interference were used for quantification with an average signal-to-noise filter of >10. The data was filtered to retain Master proteins that were measured in at least 50 mice.
HMDP heart mRNA and miRNA expression analysis
Using the miRNeasy Mini Kit (QIAGEN), total RNA was extracted from HMDP heart tissues. From this, global mRNA expression were analyzed as previously described (Cao et al., 2022), while the QIAseq miRNA Library Kit (QIAGEN) was used to create miRNA libraries. These libraries were then sequenced using 1x50 HiSeq sequencing. Reads were mapped using hisat2 (version 2.0.6) and counted using htseq-count (version 0.13.5). Differential expression analysis was performed with DESeq2.
Association mapping
Genotypes for the mouse strains were obtained using the Mouse Diversity Array (Bennett et al., 2010). After filtering for quality or missing genotypes, about 200,000 remained. Genome-wide association for phenotypes and protein abundance levels was performed using Factored Spectrally Transformed Linear Mixed Models (FaST-LMM), which applies a linear mixed model to correct for population structure (Lippert et al., 2011). A cutoff value for genome-wide significance was set at 4.1E-06, as determined previously (Bennett et al., 2010).
Cell culture and treatments
Following isolation, neonatal rat ventricular myocytes (NRVMs) were plated in DMEM containing 10% Fetal bovine serum (FBS) and 1% antibiotics overnight. The next day, NRVMs were changed to serum-free medium in the presence or absence of 100 µM phenylephrine (Sigma, Cat# P6126-10G). The cells were then transfected with miRNA control, mature miR-27b-5p, or miR-27b-3p mimics (Sigma) for 48 hr. The cell lysates were harvested for immunoblotting using primary antibodies against NDUFS4 (# sc-100567, Santa Cruz) and Actin (# 8457 S, Cell Signaling). Band densitometry was quantified using ImageJ Gel Plugin (NIH).
| Reagent | Mature sequence 5’ to 3’ |
|---|---|
| MISSION microRNA, Negative Control 1 (miRNA control) |
GGUUCGUACGUACACUGUUCA |
| MISSION microRNA - hsa-miR-27b* (miR-27b-5p) |
AGAGCUUAGCUGAUUGGUGAAC |
| MISSION microRNA - hsa-miR-27b (miR-27b-3p) |
UUCACAGUGGCUAAGUUCUGC |
Mitochondria isolation from frozen hearts
Twenty mg of frozen hearts were thawed in 1.4 mL of ice-cold isolation buffer (70 mM sucrose, 220 mM mannitol, 1 mM EGTA, 2 mM HEPES, pH 7.4 containing protease inhibitors). Hearts were mechanically homogenized with 20 strokes of a Dounce homogenizer at 4 °C. Homogenates were centrifuged at 1000 g for 10 min at 4ﹾC. Supernatant was collected (300 µL were reserved as whole lysate) and centrifuged at 10,000 g for 10 min at 4°C to obtain a pellet containing the mitochondria. Mitochondrial pellet was re-suspended in 1 mL of isolation buffer and re-centrifuged at 10,000 g for 10 min at 4°C. The mitochondrial pellet was finally re-suspended in 200 µL of isolation buffer and protein concentration determined using the BCA assay.
CoQ extraction from whole lysate and mitochondrial enriched fractions
CoQ extraction, detection and analysis was performed as described previously (Burger et al., 2020). Briefly, 15 µg protein from mitochondrial fractions or 200 µg protein from total lysate were aliquoted into 2 mL Eppendorf tubes and volumes adjusted to 100 µL for mitochondrial fractions or 200 µL for whole lysates, and samples were kept on ice for the entire process. 20 µL of 0.1 ng/mL CoQ8 (2 ng total; Avanti Polar Lipids) were added to each sample as an internal standard. To protein aliquots ice-cold 250 µL acidified methanol (0.1% HCl in MeOH) were added to each sample, followed by 300 µL hexane and samples were thoroughly mixed by vertexing. Hexane and MeOH/water layers were separated by centrifugation at 15,000 g for 5 min at 4°C. The upper layer of hexane was collected and transferred to clean 2 mL Eppendorf tubes and completely dried in a GeneVac vacuum centrifuge, on a low BP point method for 40 min (20 min pre-final stage and 20 min final stage). Samples were reconstituted in 100 µL ethanol before analysis of CoQ levels by LC-MS.
LC-MS analysis of CoQ
CoQ8, 9 and 10 levels were analyzed using a TSQ Altis triple quadrupole mass spectrometer (ThermoFisher) coupled to a Vanquish LC system (ThermoFisher). Fifteen µL of sample was injected and separated on a 2.6 μm Kinetex XB-C18 100 A column (50×2.10 mm; Phenomenex) at 45°C. Mobile Phase A consisted of 2.5 mM ammonium formate in 95% MeOH, 5% IPA and Mobile Phase B consisted of 2.4 mm ammonium formate in 100% IPA. A gradient method over 5 min was used with an initial concentration of 0% B held for 1 min before increased to 45% B over 1 min and held for 1 min, before decreasing back to 0% B over 0.5 min and column re-equilibrated over 1.5 min. Eluent was then directed into the QQQ with the following settings: source voltage = 3500 V; sheath gas 2; aux gas 2; transfer capillary temperature = 350 ﹾC. Ammonium adducts of each of the analytes were detected by SRM with Q1 and Q3 resolution set to 0.7 FWHM with the following parameters: [CoQ8 +NH4]+, m/z 744.9 → 197.1 with collision energy 32.76; [CoQ9 +NH4]+, m/z 812.9 → 197.1 with collision energy 32.76; [CoQ9H2+NH4]+, m/z 814.9 → 197.1 with collision energy 36.4; [CoQ10 +NH4]+, m/z 880.9 → 197.1 with collision energy 32.76; and [CoQ10H2+NH4]+, m/z 882.9 → 197.1 with collision energy 36.4. CoQ9 and CoQ10 were quantified in samples against a standard curve (0–1,000 nM) and normalized to spiked in CoQ8 levels (20 ng/mL) in each sample and standard. No CoQ9H2 and CoQ10H2 were detected in any of the samples, though some bleed through into these channels was detected for the oxidized analytes, though these are resolved from the reduced standard peaks (as reported previously).
Statistical analysis
Statistical analyses were performed using Prism v9.4.0 (GraphPad Software, Inc, La Jolla, CA, USA). Errors bars plotted on graphs are presented as the mean ± SEM unless reported otherwise. The critical significance value (α) was set at 0.05, and if the p values were less than α, we reported that the observed differences were statistically significant. Correlations were calculated using biweight midcorrelation using the bicorAndPvalue function of the WGCNA package (Langfelder and Horvath, 2008).
Acknowledgements
This work was supported by NIH grants DK120342, HL148577 and HL147883 (AJL) and DOD grant W81XWH2110115 (AJL); R00DK120875 (KCK); R00HL138193 (MMS); R01DK101573, R01DK102948, and RC2DK125961 (ADA); Wisconsin Alumni Research Foundation (MPK); National Health and Medical Research Council of Australia (NHMRC) grants and fellowships (DEJ and BLP); Systems Biology Association fellowship, Foundation Sorbonne fellowship, French Minister, and Master BIP (EJEH). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Karthickeyan Chella Krishnan, Email: chellakn@ucmail.uc.edu.
Aldons J Lusis, Email: JLusis@mednet.ucla.edu.
Hossein Ardehali, Northwestern University, United States.
Matthias Barton, University of Zurich, Switzerland.
Funding Information
This paper was supported by the following grants:
National Institutes of Health DK120342 to Aldons J Lusis.
National Institutes of Health R00DK120875 to Karthickeyan Chella Krishnan.
National Institutes of Health R00HL138193 to Marcus M Seldin.
National Institutes of Health R01DK101573 to Alan D Attie.
U.S. Department of Defense W81XWH2110115 to Aldons J Lusis.
Wisconsin Alumni Research Foundation to Mark P Keller.
National Health and Medical Research Council to Benjamin Parker.
National Institutes of Health R01DK102948 to Alan D Attie.
National Institutes of Health RC2DK125961 to Alan D Attie.
National Institutes of Health HL148577 to Aldons J Lusis.
National Institutes of Health HL147883 to Aldons J Lusis.
Systems Biology Association fellowship to Elie-Julien El Hachem.
Foundation Sorbonne fellowship to Elie-Julien El Hachem.
French Minister and Master BIP to Elie-Julien El Hachem.
Additional information
Competing interests
No competing interests declared.
No competing interests declared.
Senior Editor, eLife; Senior Editor for the Systems Genetics Special Issue (DEJ had no involvement in the decision making for this submission).
Author contributions
Conceptualization, Data curation, Formal analysis, Supervision, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing - original draft, Project administration, Writing – review and editing.
Formal analysis, Investigation, Visualization.
Formal analysis, Investigation, Visualization, Methodology.
Formal analysis, Investigation, Methodology.
Formal analysis, Investigation, Visualization, Methodology.
Formal analysis, Investigation, Visualization, Methodology.
Formal analysis, Investigation.
Formal analysis, Investigation.
Formal analysis, Investigation, Visualization.
Formal analysis, Investigation, Visualization.
Formal analysis, Investigation, Visualization, Methodology.
Formal analysis, Investigation.
Formal analysis, Investigation.
Resources, Supervision, Funding acquisition.
Resources, Supervision, Funding acquisition.
Formal analysis, Investigation, Visualization.
Resources, Supervision, Funding acquisition.
Resources, Supervision, Funding acquisition.
Resources, Supervision, Funding acquisition.
Formal analysis, Investigation, Visualization, Methodology, Writing – review and editing.
Resources, Supervision, Funding acquisition, Writing – review and editing.
Conceptualization, Resources, Supervision, Funding acquisition, Project administration, Writing – review and editing.
Ethics
Mice.All mice were purchased from The Jackson Laboratory and bred at UCLA according to approved institutional animal care and use committee (IACUC) protocols with daily monitoring. All mice were maintained on a 14 h light/10 h dark cycle (light is on between 6 a.m. and 8 p.m.) at a temperature of 25 degrees and 30-70% humidity. On the day of the experiment, the mice were sacrificed after 4-hour fasting.Studies in Diversity Outbred mice.All experiments that used Diversity Outbred (DO) mice were conducted in the Biochemistry Department at the University of Wisconsin-Madison and approved by the university's Animal Care and Use Committee. Briefly, 4-week-old DO mice were purchased from the Jackson Labs (stock no. 009376) and placed on a Western-style diet high in fat and sucrose (17.3% protein, 34% carbohydrate, and 44.6% kcal fat) from Envigo Teklad (TD.08811).
Additional files
(a) List of HMDP strains used for proteomic analyses and their respective genotypes at the three loci. (b) Five modules of WGCNA and their corresponding proteins. (c) List of proteins associated with chr13 locus listed in Figure 2A. The associated P values between the respective proteins and chr13 locus eigengene pSNP are listed. (d) List of proteins associated with chr17 locus listed in Figure 3A. The associated P values between the respective proteins and chr17 locus eigengene pSNP are listed. (e) List of proteins associated with chr7 locus listed in Figure 4A. The associated P values between the respective proteins and chr7 locus eigengene pSNP are listed. (f) Scoring of miRNA targeting Ndufs4 by nine algorithms and one dataset of experimental validation. (g) List of differentially abundant proteins between the control and Ndufs4-cKO groups, and their P values shown in Figure 2. Only proteins that passed the signifcant cut-off (abs[log2FC]>1 and Padj <0.001) are listed. Proteins overlapping chr13 locus are shown in blue font. (h) List of 157 mt-eQTL (LOD >6) across five tissues in the DO mice as shown in Figure 4A. (i) SNP association profile for mt-Nd5 in heart as shown in Figure 4C. (j) Protein-protein correlations between the three candidates and known hypertrophic markers. The bicor P values between the respective markers and each locus candidate gene are listed.
Data availability
All sequencing raw data can be accessed at the Gene Expression Omnibus under accession GSE194198 (HF/HS heart HMDP RNA-seq data) and GSE207142 (HF/HS heart HMDP miRNA-seq data). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE Perez-Riverol et al., 2022 partner repository with the dataset identifiers PXD036398 (HMDP hearts) and PXD036399 (NDUFS4 KO hearts).
The following datasets were generated:
Lusis AJ, Chella Krishnan K, Kaczor-Urbanowicz KE. 2022. Small RNA sequencing of hearts from female mice on high fat diet, from 85 strains. NCBI Gene Expression Omnibus. GSE207142
Lusis AJ, Chella Krishnan K, Parker BL. 2022. HMDP hearts. ProteomeXchange. PXD036398
Lusis AJ, Chella Krishnan K, Parker BL. 2022. NDUFS4 KO hearts. ProteomeXchange. PXD036399
The following previously published dataset was used:
Lusis AJ, Cao Y. 2022. Heart profiling in HMDP (high-fat/high-sucrose diet) NCBI Gene Expression Omnibus. GSE194198
References
- Adjobo-Hermans MJW, de Haas R, Willems P, Wojtala A, van Emst-de Vries SE, Wagenaars JA, van den Brand M, Rodenburg RJ, Smeitink JAM, Nijtmans LG, Sazanov LA, Wieckowski MR, Koopman WJH. Ndufs4 deletion triggers loss of Ndufa12 in Ndufs4(-/-) mice and Leigh syndrome patients: A stabilizing role for Ndufaf2. Biochimica et Biophysica Acta. Bioenergetics. 2020;1861:148213. doi: 10.1016/j.bbabio.2020.148213. [DOI] [PubMed] [Google Scholar]
- Aguet F. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AT, Boehme L, Carbajosa G, Seitan VC, Small KS, Hodgkinson A. Nuclear genetic regulation of the human mitochondrial transcriptome. eLife. 2019;8:e41927. doi: 10.7554/eLife.41927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begriche K, Igoudjil A, Pessayre D, Fromenty B. Mitochondrial dysfunction in NASH: causes, consequences and possible means to prevent it. Mitochondrion. 2006;6:1–28. doi: 10.1016/j.mito.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang W, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ. A high-resolution Association mapping panel for the dissection of complex traits in mice. Genome Research. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, Perry JB, Allen ME, Sabbah HN, Stauffer BL, Shaikh SR, Cleland JGF, Colucci WS, Butler J, Voors AA, Anker SD, Pitt B, Pieske B, Filippatos G, Greene SJ, Gheorghiade M. Expert consensus document: mitochondrial function as a therapeutic target in heart failure. Nature Reviews Cardiology. 2017;14:238–250. doi: 10.1038/nrcardio.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynedal B, Choi J, Raj T, Bjornson R, Stranger BE, Neale BM, Voight BF, Cotsapas C. Large-scale Trans-eQTLs affect hundreds of transcripts and mediate patterns of transcriptional Co-regulation. American Journal of Human Genetics. 2017;100:581–591. doi: 10.1016/j.ajhg.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger N, Logan A, Prime TA, Mottahedin A, Caldwell ST, Krieg T, Hartley RC, James AM, Murphy MP. A sensitive mass spectrometric assay for mitochondrial Coq pool redox state in vivo. Free Radical Biology & Medicine. 2020;147:37–47. doi: 10.1016/j.freeradbiomed.2019.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Vergnes L, Wang YC, Pan C, Chella Krishnan K, Moore TM, Rosa-Garrido M, Kimball TH, Zhou Z, Charugundla S, Rau CD, Seldin MM, Wang J, Wang Y, Vondriska TM, Reue K, Lusis AJ. Sex differences in heart mitochondria regulate diastolic dysfunction. Nature Communications. 2022;13:3850. doi: 10.1038/s41467-022-31544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chella Krishnan K, Kurt Z, Barrere-Cain R, Sabir S, Das A, Floyd R, Vergnes L, Zhao Y, Che N, Charugundla S, Qi H, Zhou Z, Meng Y, Pan C, Seldin MM, Norheim F, Hui S, Reue K, Lusis AJ, Yang X. Integration of multi-Omics data from mouse diversity panel highlights mitochondrial dysfunction in non-alcoholic fatty liver disease. Cell Systems. 2018;6:103–115. doi: 10.1016/j.cels.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chella Krishnan K, Sabir S, Shum M, Meng Y, Acín-Pérez R, Lang JM, Floyd RR, Vergnes L, Seldin MM, Fuqua BK, Jayasekera DW, Nand SK, Anum DC, Pan C, Stiles L, Péterfy M, Reue K, Liesa M, Lusis AJ. Sex-specific metabolic functions of Adipose Lipocalin-2. Molecular Metabolism. 2019;30:30–47. doi: 10.1016/j.molmet.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chella Krishnan K, Vergnes L, Acín-Pérez R, Stiles L, Shum M, Ma L, Mouisel E, Pan C, Moore TM, Péterfy M, Romanoski CE, Reue K, Björkegren JLM, Laakso M, Liesa M, Lusis AJ. Sex-specific genetic regulation of Adipose mitochondria and metabolic syndrome by Ndufv2. Nature Metabolism. 2021;3:1552–1568. doi: 10.1038/s42255-021-00481-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Methner C, Buonincontri G, Hu C-H, Logan A, Sawiak SJ, Murphy MP, Krieg T. Complex I deficiency due to selective loss of Ndufs4 in the mouse heart results in severe hypertrophic cardiomyopathy. PLOS ONE. 2014;9:e94157. doi: 10.1371/journal.pone.0094157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Ohira T, Sakaguchi Y, Goshima N, Nomura N, Nagao A, Suzuki T. LRPPRC/SLIRP suppresses Pnpase-mediated mRNA decay and promotes Polyadenylation in human mitochondria. Nucleic Acids Research. 2012;40:8033–8047. doi: 10.1093/nar/gks506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems Genetics approaches to understand complex traits. Nature Reviews. Genetics. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate Tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkersen L, Gustafsson S, Wang Q, Hansen DH, Hedman ÅK, Schork A, Page K, Zhernakova DV, Wu Y, Peters J, Eriksson N, Bergen SE, Boutin TS, Bretherick AD, Enroth S, Kalnapenkis A, Gådin JR, Suur BE, Chen Y, Matic L, Gale JD, Lee J, Zhang W, Quazi A, Ala-Korpela M, Choi SH, Claringbould A, Danesh J, Davey Smith G, de Masi F, Elmståhl S, Engström G, Fauman E, Fernandez C, Franke L, Franks PW, Giedraitis V, Haley C, Hamsten A, Ingason A, Johansson Å, Joshi PK, Lind L, Lindgren CM, Lubitz S, Palmer T, Macdonald-Dunlop E, Magnusson M, Melander O, Michaelsson K, Morris AP, Mägi R, Nagle MW, Nilsson PM, Nilsson J, Orho-Melander M, Polasek O, Prins B, Pålsson E, Qi T, Sjögren M, Sundström J, Surendran P, Võsa U, Werge T, Wernersson R, Westra H-J, Yang J, Zhernakova A, Ärnlöv J, Fu J, Smith JG, Esko T, Hayward C, Gyllensten U, Landen M, Siegbahn A, Wilson JF, Wallentin L, Butterworth AS, Holmes MV, Ingelsson E, Mälarstig A. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nature Metabolism. 2020;2:1135–1148. doi: 10.1038/s42255-020-00287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium The Gtex consortium Atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Wu M, Guo R, Yan K, Lei J, Gao N, Yang M. The architecture of the mammalian Respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- He B, Shi J, Wang X, Jiang H, Zhu HJ. Genome-wide pQTL analysis of protein expression regulatory networks in the human liver. BMC Biology. 2020;18:97. doi: 10.1186/s12915-020-00830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J. Mitochondrial complex I. Annual Review of Biochemistry. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large Gene lists using DAVID Bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hui ST, Kurt Z, Tuominen I, Norheim F, Davis RC, Pan C, Dirks DL, Magyar CE, French SW, Chella Krishnan K, Sabir S, Campos-Pérez F, Méndez-Sánchez N, Macías-Kauffer L, León-Mimila P, Canizales-Quinteros S, Yang X, Beaven SW, Huertas-Vazquez A, Lusis AJ. The genetic architecture of diet-induced hepatic fibrosis in mice. Hepatology. 2018;68:2182–2196. doi: 10.1002/hep.30113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlhöfer F, Kmita K, Wittig I, Zwicker K, Zickermann V. Accessory subunit NUYM (Ndufs4) is required for stability of the electron input Module and activity of mitochondrial complex I. Biochimica et Biophysica Acta - Bioenergetics. 2017;1858:175–181. doi: 10.1016/j.bbabio.2016.11.010. [DOI] [PubMed] [Google Scholar]
- Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nature Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Käll L, Storey JD, Noble WS. QVALITY: non-parametric estimation of Q-values and posterior error probabilities. Bioinformatics. 2009;25:964–966. doi: 10.1093/bioinformatics/btp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanlidis G, Lee CF, Garcia-Menendez L, Kolwicz SC, Suthammarak W, Gong G, Sedensky MM, Morgan PG, Wang W, Tian R. Mitochondrial complex I deficiency increases protein Acetylation and accelerates heart failure. Cell Metabolism. 2013;18:239–250. doi: 10.1016/j.cmet.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MP, Paul PK, Rabaglia ME, Stapleton DS, Schueler KL, Broman AT, Ye SI, Leng N, Brandon CJ, Neto EC, Plaisier CL, Simonett SP, Kebede MA, Sheynkman GM, Klein MA, Baliga NS, Smith LM, Broman KW, Yandell BS, Kendziorski C, Attie AD. The transcription factor Nfatc2 regulates beta-cell proliferation and genes associated with type 2 diabetes in mouse and human Islets. PLOS Genetics. 2016;12:e1006466. doi: 10.1371/journal.pgen.1006466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MP, Gatti DM, Schueler KL, Rabaglia ME, Stapleton DS, Simecek P, Vincent M, Allen S, Broman AT, Bacher R, Kendziorski C, Broman KW, Yandell BS, Churchill GA, Attie AD. Genetic drivers of Pancreatic Islet function. Genetics. 2018;209:335–356. doi: 10.1534/genetics.118.300864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MP, Rabaglia ME, Schueler KL, Stapleton DS, Gatti DM, Vincent M, Mitok KA, Wang Z, Ishimura T, Simonett SP, Emfinger CH, Das R, Beck T, Kendziorski C, Broman KW, Yandell BS, Churchill GA, Attie AD. Gene Loci associated with insulin secretion in Islets from non-diabetic mice. The Journal of Clinical Investigation. 2019;129:4419–4432. doi: 10.1172/JCI129143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circulation Research. 2008;102:401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Yu J, Bi J, Qi H, Di W, Wu L, Wang L, Zha J, Lv S, Zhang F, Li Y, Hu F, Liu F, Zhou H, Liu J, Ding G. Glucocorticoids Transcriptionally regulate miR-27B expression promoting body fat accumulation via suppressing the Browning of white adipose tissue. Diabetes. 2015;64:393–404. doi: 10.2337/db14-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA Annotation and deep-sequencing data. Nucleic Acids Research. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/- mice. The Journal of Biological Chemistry. 2008;283:26217–26227. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Stepanyan Z, Bigras E, Hekimi S. Reversal of the mitochondrial phenotype and slow development of oxidative biomarkers of aging in long-lived Mclk1+/- mice. The Journal of Biological Chemistry. 2009;284:20364–20374. doi: 10.1074/jbc.M109.006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Wang Y, Bigras E, Hekimi S. The Submitochondrial distribution of Ubiquinone affects respiration in long-lived Mclk1+/- mice. The Journal of Cell Biology. 2012;199:215–224. doi: 10.1083/jcb.201203090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong DW, Komen JC, Hewitt CA, Arnaud E, McKenzie M, Phipson B, Bahlo M, Laskowski A, Kinkel SA, Davey GM, Heath WR, Voss AK, Zahedi RP, Pitt JJ, Chrast R, Sickmann A, Ryan MT, Smyth GK, Thorburn DR, Scott HS. Proteomic and Metabolomic analyses of mitochondrial complex I-deficient mouse model generated by spontaneous B2 short interspersed nuclear element (SINE) insertion into NADH dehydrogenase (Ubiquinone) Fe-S protein 4 (Ndufs4) Gene. The Journal of Biological Chemistry. 2012;287:20652–20663. doi: 10.1074/jbc.M111.327601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshinsky-Silver E, Lebre A-S, Minai L, Saada A, Steffann J, Cohen S, Rötig A, Munnich A, Lev D, Lerman-Sagie T. Ndufs4 mutations cause Leigh syndrome with predominant brainstem involvement. Molecular Genetics and Metabolism. 2009;97:185–189. doi: 10.1016/j.ymgme.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Li G, Shao Y, Guo HC, Zhi Y, Qiao B, Ma K, Du J, Lai YQ, Li Y. Microrna-27B-3P down-regulates Fgf1 and aggravates pathological cardiac remodelling. Cardiovascular Research. 2022;118:2139–2151. doi: 10.1093/cvr/cvab248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. Fast linear mixed models for genome-wide Association studies. Nature Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the Clk-1-dependent mechanism of longevity: loss of Mclk1 increases cellular fitness and lifespan in mice. Genes & Development. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis AJ, Seldin MM, Allayee H, Bennett BJ, Civelek M, Davis RC, Eskin E, Farber CR, Hui S, Mehrabian M, Norheim F, Pan C, Parks B, Rau CD, Smith DJ, Vallim T, Wang Y, Wang J. The hybrid mouse diversity panel: a resource for systems Genetics analyses of metabolic and cardiovascular traits. Journal of Lipid Research. 2016;57:925–942. doi: 10.1194/jlr.R066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. Multinotch Ms3 enables accurate, sensitive, and Multiplexed detection of differential expression across cancer cell line Proteomes. Analytical Chemistry. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, Mitchell GA, Morin C, Mann M, Hudson TJ, Robinson B, Rioux JD, Lander ES. Identification of a gene causing human cytochrome C oxidase deficiency by integrative Genomics. PNAS. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AP, Fu CP, Kao CY, Welsh CE, Didion JP, Yadgary L, Hyacinth L, Ferris MT, Bell TA, Miller DR, Giusti-Rodriguez P, Nonneman RJ, Cook KD, Whitmire JK, Gralinski LE, Keller M, Attie AD, Churchill GA, Petkov P, Sullivan PF, Brennan JR, McMillan L, Pardo-Manuel de Villena F. The mouse universal Genotyping array: from Substrains to subspecies. G3: Genes, Genomes, Genetics. 2015;6:263–279. doi: 10.1534/g3.115.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Ardehali H, Balaban RS, DiLisa F, Dorn GW, Kitsis RN, Otsu K, Ping P, Rizzuto R, Sack MN, Wallace D, Youle RJ, American Heart Association Council on Basic Cardiovascular Sciences, Council on Clinical Cardiology, and Council on Functional Genomics and Translational Biology Mitochondrial function, biology, and role in disease: A scientific statement from the American heart Association. Circulation Research. 2016;118:1960–1991. doi: 10.1161/RES.0000000000000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai D, Yuasa S, Takahashi M, Shimizu T, Asaumi S, Isono K, Takao T, Suzuki Y, Kuroyanagi H, Hirokawa K, Koseki H, Shirsawa T. Mouse Homologue of Coq7/Clk-1, longevity Gene in Caenorhabditis elegans, is essential for coenzyme Q synthesis, maintenance of mitochondrial integrity, and neurogenesis. Biochemical and Biophysical Research Communications. 2001;289:463–471. doi: 10.1006/bbrc.2001.5977. [DOI] [PubMed] [Google Scholar]
- Norheim F, Hui ST, Kulahcioglu E, Mehrabian M, Cantor RM, Pan C, Parks BW, Lusis AJ. Genetic and hormonal control of hepatic steatosis in female and male mice. Journal of Lipid Research. 2017;58:178–187. doi: 10.1194/jlr.M071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norheim F, Bjellaas T, Hui ST, Chella Krishnan K, Lee J, Gupta S, Pan C, Hasin-Brumshtein Y, Parks BW, Li DY, Bui HH, Mosier M, Wu Y, Huertas-Vazquez A, Hazen SL, Gundersen TE, Mehrabian M, Tang WHW, Hevener AL, Drevon CA, Lusis AJ. Genetic, dietary, and sex-specific regulation of hepatic Ceramides and the relationship between hepatic Ceramides and IR. Journal of Lipid Research. 2018;59:1164–1174. doi: 10.1194/jlr.M081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norheim F, Hasin-Brumshtein Y, Vergnes L, Chella Krishnan K, Pan C, Seldin MM, Hui ST, Mehrabian M, Zhou Z, Gupta S, Parks BW, Walch A, Reue K, Hofmann SM, Arnold AP, Lusis AJ. Gene-by-sex interactions in mitochondrial functions and Cardio-metabolic traits. Cell Metabolism. 2019;29:932–949. doi: 10.1016/j.cmet.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oláhová M, Hardy SA, Hall J, Yarham JW, Haack TB, Wilson WC, Alston CL, He L, Aznauryan E, Brown RM, Brown GK, Morris AAM, Mundy H, Broomfield A, Barbosa IA, Simpson MA, Deshpande C, Moeslinger D, Koch J, Stettner GM, Bonnen PE, Prokisch H, Lightowlers RN, McFarland R, Chrzanowska-Lightowlers ZMA, Taylor RW. LRPPRC mutations cause early-onset Multisystem mitochondrial disease outside of the French-Canadian population. Brain. 2015;138:3503–3519. doi: 10.1093/brain/awv291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, Lusis AJ. Sex differences and hormonal effects on gut Microbiota composition in mice. Gut Microbes. 2016;7:313–322. doi: 10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, Mehrabian M, Ursell LK, He A, Castellani LW, Zinker B, Kirby M, Drake TA, Drevon CA, Knight R, Gargalovic P, Kirchgessner T, Eskin E, Lusis AJ. Genetic control of obesity and gut Microbiota composition in response to high-fat, high-Sucrose diet in mice. Cell Metabolism. 2013;17:141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Sallam T, Mehrabian M, Psychogios N, Hui ST, Norheim F, Castellani LW, Rau CD, Pan C, Phun J, Zhou Z, Yang W-P, Neuhaus I, Gargalovic PS, Kirchgessner TG, Graham M, Lee R, Tontonoz P, Gerszten RE, Hevener AL, Lusis AJ. Genetic architecture of insulin resistance in the mouse. Cell Metabolism. 2015;21:334–347. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, Walzer M, Wang S, Brazma A, Vizcaíno JA. The PRIDE database resources in 2022: a Hub for mass Spectrometry-based Proteomics evidences. Nucleic Acids Research. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S, Sharma R, Gupta R, Ast T, Chan C, Durham TJ, Goodman RP, Grabarek Z, Haas ME, Hung WHW, Joshi PR, Jourdain AA, Kim SH, Kotrys AV, Lam SS, McCoy JG, Meisel JD, Miranda M, Panda A, Patgiri A, Rogers R, Sadre S, Shah H, Skinner OS, To T-L, Walker MA, Wang H, Ward PS, Wengrod J, Yuan C-C, Calvo SE, Mootha VK. Mitocarta3.0: an updated mitochondrial Proteome now with sub-Organelle localization and pathway annotations. Nucleic Acids Research. 2021;49:D1541–D1547. doi: 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau CD, Wang J, Avetisyan R, Romay MC, Martin L, Ren S, Wang Y, Lusis AJ. Mapping genetic contributions to cardiac pathology induced by beta-adrenergic stimulation in mice. Circulation. Cardiovascular Genetics. 2015;8:40–49. doi: 10.1161/CIRCGENETICS.113.000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau CD, Romay MC, Tuteryan M, Wang JJC, Santolini M, Ren S, Karma A, Weiss JN, Wang Y, Lusis AJ. Systems Genetics approach identifies Gene pathways and Adamts2 as drivers of isoproterenol-induced cardiac hypertrophy and cardiomyopathy in mice. Cell Systems. 2017;4:121–128. doi: 10.1016/j.cels.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffieux H, Carayol J, Popescu R, Harper ME, Dent R, Saris WHM, Astrup A, Hager J, Davison AC, Valsesia A. A fully joint Bayesian quantitative trait locus mapping of human protein abundance in plasma. PLOS Computational Biology. 2020;16:e1007882. doi: 10.1371/journal.pcbi.1007882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Cámara Y, Milenkovic D, Zickermann V, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, Brandt U, Stewart JB, Gustafsson CM, Larsson N-G. LRPPRC is necessary for Polyadenylation and coordination of translation of mitochondrial mRNAs. The EMBO Journal. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal AJ, Campbell–Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, Luketic VA, Shiffman ML, Clore JN. Nonalcoholic Steatohepatitis: Association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- Scacco S, Petruzzella V, Budde S, Vergari R, Tamborra R, Panelli D, van den Heuvel LP, Smeitink JA, Papa S. Pathological mutations of the human Ndufs4 Gene of the 18-kDa (AQDQ) subunit of complex I affect the expression of the protein and the assembly and function of the complex. The Journal of Biological Chemistry. 2003;278:44161–44167. doi: 10.1074/jbc.M307615200. [DOI] [PubMed] [Google Scholar]
- Seldin MM, Kim ED, Romay MC, Li S, Rau CD, Wang JJ, Krishnan KC, Wang Y, Deb A, Lusis AJ. A systems Genetics approach identifies Trp53Inp2 as a link between cardiomyocyte glucose utilization and hypertrophic response. American Journal of Physiology. Heart and Circulatory Physiology. 2017;312:H728–H741. doi: 10.1152/ajpheart.00068.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MM, Koplev S, Rajbhandari P, Vergnes L, Rosenberg GM, Meng Y, Pan C, Phuong TMN, Gharakhanian R, Che N, Mäkinen S, Shih DM, Civelek M, Parks BW, Kim ED, Norheim F, Chella Krishnan K, Hasin-Brumshtein Y, Mehrabian M, Laakso M, Drevon CA, Koistinen HA, Tontonoz P, Reue K, Cantor RM, Björkegren JLM, Lusis AJ. A strategy for discovery of endocrine interactions with application to whole-body metabolism. Cell Metabolism. 2018;27:1138–1155. doi: 10.1016/j.cmet.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin M, Yang X, Lusis AJ. Systems Genetics applications in metabolism research. Nature Metabolism. 2019;1:1038–1050. doi: 10.1038/s42255-019-0132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Fonarow GC, Butler J, Ezekowitz JA, Felker GM. Coenzyme Q10 and heart failure: A state-of-the-art review. Circulation. Heart Failure. 2016;9:e002639. doi: 10.1161/CIRCHEARTFAILURE.115.002639. [DOI] [PubMed] [Google Scholar]
- Siira SJ, Spåhr H, Shearwood A-MJ, Ruzzenente B, Larsson N-G, Rackham O, Filipovska A. LRPPRC-mediated folding of the mitochondrial transcriptome. Nature Communications. 2017;8:1532. doi: 10.1038/s41467-017-01221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small KS, Todorčević M, Civelek M, El-Sayed Moustafa JS, Wang X, Simon MM, Fernandez-Tajes J, Mahajan A, Horikoshi M, Hugill A, Glastonbury CA, Quaye L, Neville MJ, Sethi S, Yon M, Pan C, Che N, Viñuela A, Tsai P-C, Nag A, Buil A, Thorleifsson G, Raghavan A, Ding Q, Morris AP, Bell JT, Thorsteinsdottir U, Stefansson K, Laakso M, Dahlman I, Arner P, Gloyn AL, Musunuru K, Lusis AJ, Cox RD, Karpe F, McCarthy MI. Regulatory variants at Klf14 influence type 2 diabetes risk via a female-specific effect on Adipocyte size and body composition. Nature Genetics. 2018;50:572–580. doi: 10.1038/s41588-018-0088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, Osellame LD, Stait T, Beilharz TH, Thorburn DR, Salim A, Ryan MT. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- Sun L, Trajkovski M. Mir-27 Orchestrates the transcriptional regulation of Brown Adipogenesis. Metabolism. 2014;63:272–282. doi: 10.1016/j.metabol.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. High-resolution genetic mapping using the mouse diversity Outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial genetic medicine. Nature Genetics. 2018;50:1642–1649. doi: 10.1038/s41588-018-0264-z. [DOI] [PubMed] [Google Scholar]
- Wang J, Song Y, Zhang Y, Xiao H, Sun Q, Hou N, Guo S, Wang Y, Fan K, Zhan D, Zha L, Cao Y, Li Z, Cheng X, Zhang Y, Yang X. Cardiomyocyte overexpression of miR-27B induces cardiac hypertrophy and dysfunction in mice. Cell Research. 2012;22:516–527. doi: 10.1038/cr.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Oxer D, Hekimi S. Mitochondrial function and LifeSpan of mice with controlled Ubiquinone biosynthesis. Nature Communications. 2015;6:6393. doi: 10.1038/ncomms7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JJC, Rau C, Avetisyan R, Ren S, Romay MC, Stolin G, Gong KW, Wang Y, Lusis AJ. Genetic dissection of cardiac remodeling in an isoproterenol-induced heart failure mouse model. PLOS Genetics. 2016;12:e1006038. doi: 10.1371/journal.pgen.1006038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cai H, Li H, Gao Z, Song K. Atrial overexpression of microRNA-27B attenuates angiotensin II-induced atrial fibrosis and fibrillation by targeting Alk5. Human Cell. 2018;31:251–260. doi: 10.1007/s13577-018-0208-z. [DOI] [PubMed] [Google Scholar]
- Yang C, Farias FHG, Ibanez L, Suhy A, Sadler B, Fernandez MV, Wang F, Bradley JL, Eiffert B, Bahena JA, Budde JP, Li Z, Dube U, Sung YJ, Mihindukulasuriya KA, Morris JC, Fagan AM, Perrin RJ, Benitez BA, Rhinn H, Harari O, Cruchaga C. Genomic Atlas of the Proteome from brain, CSF and plasma Prioritizes proteins implicated in neurological disorders. Nature Neuroscience. 2021;24:1302–1312. doi: 10.1038/s41593-021-00886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Joehanes R, Johnson AD, Huan T, Liu C, Freedman JE, Munson PJ, Hill DE, Vidal M, Levy D. Dynamic role of Trans regulation of gene expression in relation to complex traits. The American Journal of Human Genetics. 2017;100:571–580. doi: 10.1016/j.ajhg.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Chen G, Song C, Keefe J, Mendelson M, Huan T, Sun BB, Laser A, Maranville JC, Wu H, Ho JE, Courchesne P, Lyass A, Larson MG, Gieger C, Graumann J, Johnson AD, Danesh J, Runz H, Hwang S-J, Liu C, Butterworth AS, Suhre K, Levy D. Genome-wide mapping of plasma protein Qtls identifies Putatively causal genes and pathways for cardiovascular disease. Nature Communications. 2018;9:3268. doi: 10.1038/s41467-018-05512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Lanza IR, Swain JM, Sarr MG, Nair KS, Jensen MD. Adipocyte mitochondrial function is reduced in human obesity independent of fat cell size. The Journal of Clinical Endocrinology and Metabolism. 2014;99:E209–E216. doi: 10.1210/jc.2013-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Lv Y, Di W, Liu J, Kong X, Sheng Y, Huang M, Lv S, Qi H, Gao M, Liang H, Kim S, Fu Z, Zhou H, Ding G. Mir-27B-3P regulation in Browning of human visceral Adipose related to central obesity. Obesity. 2018;26:387–396. doi: 10.1002/oby.22104. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gong G, Wang P, Zhang Z, Kolwicz SC, Rabinovitch PS, Tian R, Wang W. Heart specific knockout of Ndufs4 ameliorates ischemia reperfusion injury. Journal of Molecular and Cellular Cardiology. 2018;123:38–45. doi: 10.1016/j.yjmcc.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Gummesson A, Tebani A, Karlsson MJ, Hong M-G, Schwenk JM, Edfors F, Bergström G, Fagerberg L, Uhlén M. Whole-genome sequence Association analysis of blood proteins in a longitudinal wellness cohort. Genome Medicine. 2020;12:53. doi: 10.1186/s13073-020-00755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Tian R. Mitochondrial dysfunction in pathophysiology of heart failure. Journal of Clinical Investigation. 2018;128:3716–3726. doi: 10.1172/JCI120849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]