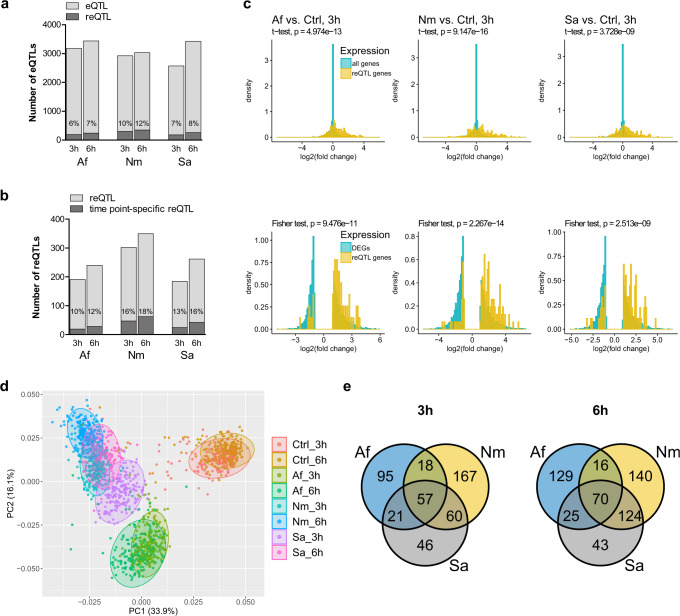
Fig. 2. Overview of reQTL effects in pathogen-exposed monocytes.
a Percentage of response expression quantitative trait loci (reQTLs) from all cis eQTLs in pathogen-exposed monocytes (A. fumigatus [Af]: 3,175 eQTLs and 191 reQTLs at 3 h, 3,439 eQTLs, and 240 reQTLs at 6 h; N. meningitidis [Nm]: 2,921 eQTLs and 302 reQTLs at 3 h, 3,029 eQTLs and 350 reQTLs at 6 h; S. aureus [Sa]: 2,574 eQTLs and 184 reQTLs at 3 h, 3,425 eQTLs, and 262 reQTLs at 6 h). b Percentage of timepoint-specific reQTLs in pathogen-exposed monocytes (Af: 19 at 3 h, 28 at 6 h; Nm: 47 at 3 h, 62 at 6 h; Sa: 24 at 3 h, 42 at 6 h). c Histograms comparing the gene expression log2FC distribution of all genes (top row) or all DEGs (|log2FC|≥ 1, bottom row) versus all reQTL-regulated genes after 3 h of stimulation (6 h stimulation: Supplementary Fig. 4a). Distribution differences were tested for statistical significance. Two-tailed t-test or Fisher exact test-based p-values are indicated. d Principal component analysis of 745 reQTL-regulated genes in monocytes of 215 healthy volunteers after exposure to Af, Nm, or Sa for 3 h and 6 h. Colors indicate stimulus and timepoint. e Venn diagrams of all reQTL-regulated genes after 3 h (left, total of 464 reQTL-regulated genes) and 6 h (right, total of 547 reQTL-regulated genes). 369 different genes are reQTL-regulated upon 3 h bacterial stimulation, 32% (117) of them are reQTL-regulated following Nm and Sa stimulation (6 h: 46% (194 overlapping reQTL-regulated genes out of 418)). 418 different genes are reQTL-regulated after 3 h stimulation with Af or Nm, 18% (75) of them are reQTL-regulated following Af and Nm stimulation (6 h: 17% (86 overlapping reQTL-regulated genes out of 504)). 297 different genes are reQTL-regulated after 3 h stimulation with Af or Sa, 26% (78) of them are reQTL-regulated following Af and Sa stimulation (6 h: 23% (95 overlapping reQTL-regulated genes out of 407)). 20% of genes are specifically reQTL-regulated after 3 h exposure to Af (95 out of 464 reQTL-regulated genes), 36% of genes are specifically reQTL-regulated after 3 h exposure to Nm (167 out of 464 reQTL-regulated genes) and 10% of genes are specifically reQTL-regulated after 3 h exposure to Sa (46 out of 464 reQTL-regulated genes) (6 h: 24% specifically reQTL-regulated genes after Af, 26% after Nm and 8% after Sa stimulation). Ctrl unstimulated, DEGs differentially expressed genes, FC fold-change. List of all DEGs is provided in Supplementary Data 1, while eQTL and reQTL data are provided in Supplementary Data 2 and 3, respectively. Source data are provided as a Source Data file.