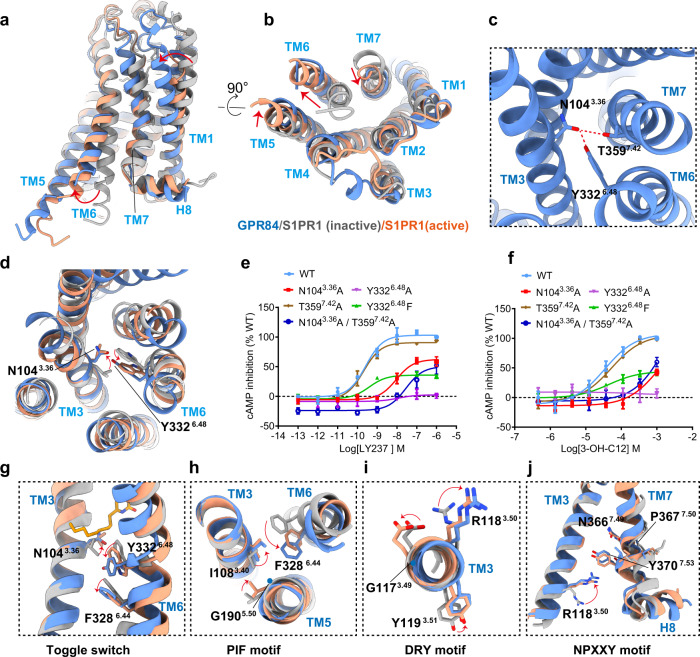
Fig. 5. GPR84 activation mechanism.
a, b Superposition of activated GPR84 (cornflower blue) with active S1PR1 (light salmon; PDB code: 7EVY) and inactive S1PR1 (gray; PDB code: 3V2Y). Notable conformational changes occur at intracellular ends of TM6 and TM7 upon receptor activation, side view (a) and top view (b). c N1043.36 forms hydrogen bond with Y3326.48 and T3597.42. d Detailed comparison of activated GPR84(cornflower blue) with inactive S1PR1 (gray; PDB code: 3V2Y) and active S1PR1 (light salmon; PDB code: 7EVY). In particular, side-chains N1043.36 and Y3326.48 exhibit similar displacement to L1283.36, W2696.48 of S1PR1 upon activation. e, f Dose-response curves of LY237(e) and 3-OH-C12(f) in activating GPR84 with mutations destroying the hydrogen bonds. Data are shown as mean ± S.E.M. from a minimum of three technical replicates, which performed in triplicates. The representative dose-response curves are shown. g The “dual toggle switch” N1043.36 and Y3324.48 of GPR84 display relative rotameric change when sensing agonist. h–j The key G-I-F6.44(PIF motif in common GPCRs) (h), G-R3.50-Y(D(E)RY motif in common GPCRs) (i), and N-P7.50-xx-Y7.53 ( j) motifs displayed conformational rearrangement in activated GPR84. Source data are provided as a Source data file.