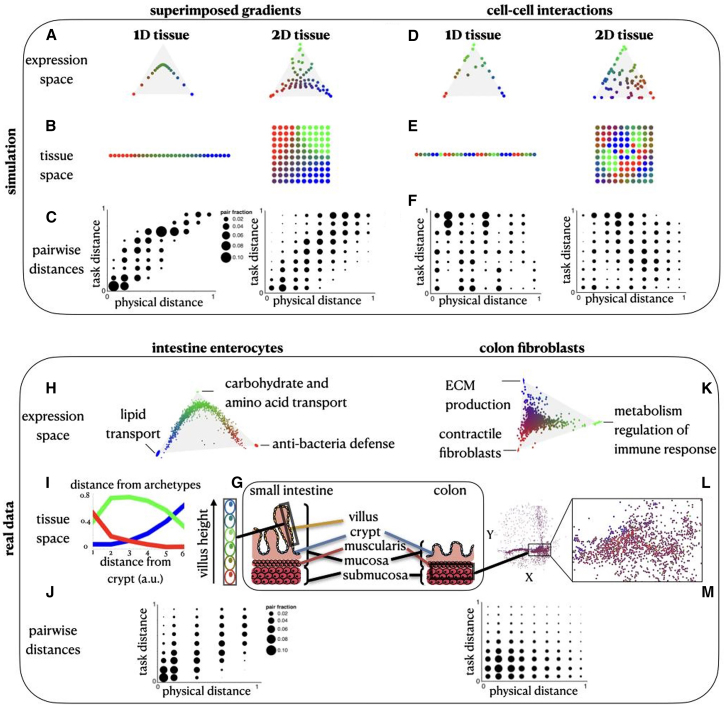
Figure 3.
Distinct spatial patterns emerge from external gradients and cell-cell interactions
(A and B) The Pareto-optimal solution of cells that are affected by an external gradient across a 1D and a 2D tissue in gene expression (A) and tissue (B) space.
(C) With external gradients, the pairwise expression distances (y axis) versus the pairwise physical distances (x axis) show high Pearson correlation (for 1D: corr = 0.88, p < 0.001, for 2D: corr = 0.72, p < 0.001).
(D and E) The Pareto-optimal solution of cells that are affected by cell-cell interactions in a 1D and a 2D tissue in gene expression (D) and tissue (E) space.
(F) With cell-cell interactions, the pairwise expression distances (y axis) versus the pairwise physical distances (x axis) are anticorrelated (in terms of Pearson correlation, for 1D, corr = −0.1, p = 0.002, for 2D, corr = −0.13, p < 0.001).
(G) Schematics of the colon tissue including the intestinal villi and the muscularis layer.
(H) Gene expression profile of enterocytes where the cells are colored by their task specializations (combinations of red, green, and blue colors representing the three tasks).
(I) Arrangements of enterocytes along the crypt-to-villus axis. The distance of the cells from each archetype is plotted as a function of the distance from the crypt.
(J) The pairwise distances in expression versus physical space of enterocytes show high Pearson correlation (corr = 0.67, p < 0.001).
(K) Gene expression profile of colon fibroblasts from Slide-seq data (Avraham-Davidi et al.24), where the cells are colored by their task specializations. At the vertices (H,K) are ellipses that indicate STD of vertex position from bootstrapping.
(L) Spatial arrangements of the fibroblasts in the colon tissue.
(M) The pairwise distances in expression versus physical space of fibroblasts show negative Pearson correlation (corr = −0.1, p < 0.001).