Figure 4.
Inferring archetype crosstalk networks of colon fibroblasts based on ligand-receptor interactions
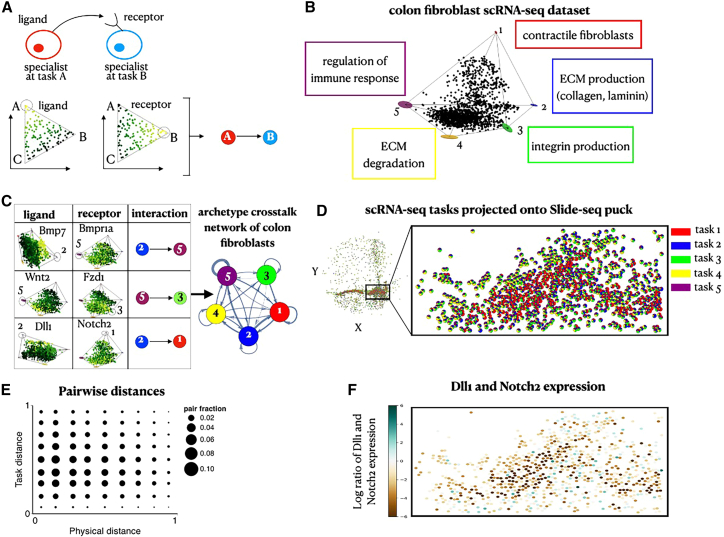
(A) Interactions between specialist cells are inferred from enrichments of ligands and their corresponding receptors. We use a directed edge to represent a pair of a ligand and its corresponding receptor that are enriched near each of the archetypes it connects.
(B) A projection of gene expression profiles of single-cell RNA sequencing (scRNA-seq) colon fibroblasts28 on the first three PCs. Fibroblasts fill in a 5-vertex polytope (p < ).
(C) A table showing examples of ligands and their respective receptors that are enriched near the archetypes. We plot the complete archetype crosstalk network inferred for the colon fibroblasts where the thickness of each edge corresponds to the number of ligand-receptor pairs enriched between its vertex archetypes.
(D and E) Using TACCO,29 we compute a mapping from fibroblast cells assayed by scRNA-seq and Slide-seq beads based on their expression agreement. Using the mapping, we (D) infer the scRNA-seq task components for each bead (depicted in pie charts per bead) and (E) compute the corresponding correlation of pairwise task distances versus physical distances (corr = −0.05, p < ).
(F) Projection of the scRNA-seq expression profiles onto the Slide-seq beads. To view the expression of Delta and Notch, we image their log ratio. Beads whose inferred Delta expression is greater/less than their Notch expression lean toward a turquoise/brown shade, respectively.