Figure 1.
Overview of scRNA-seq dataset
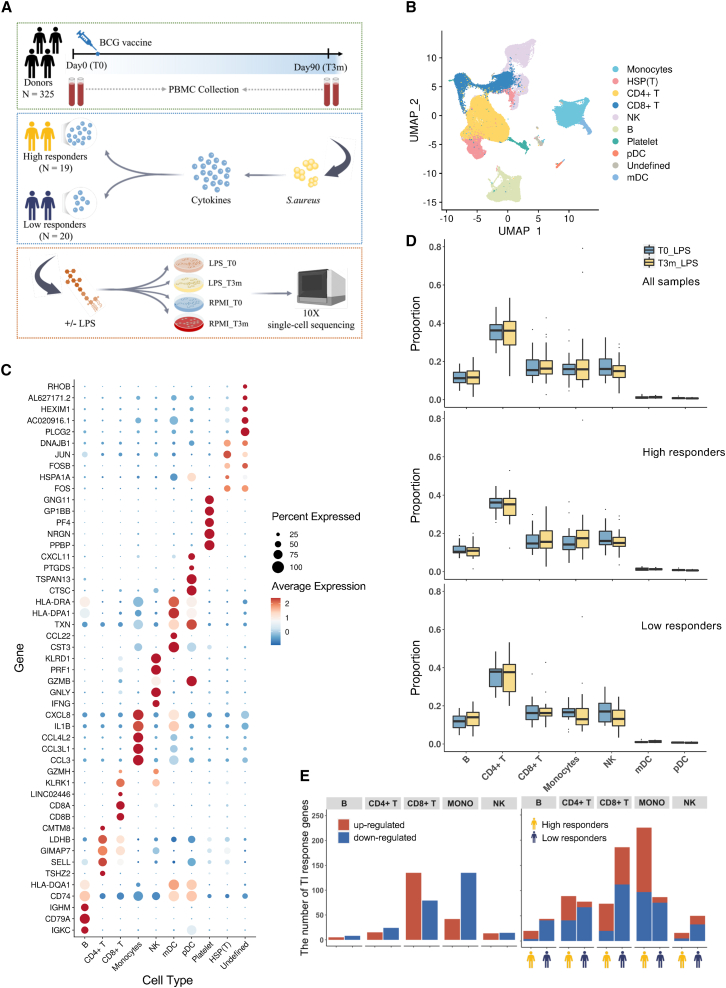
(A) Experimental design of scRNA-seq. Peripheral blood mononuclear cells (PBMCs) were collected before (T0) and 3 months after (T3m) BCG vaccination. At each time point, cells were stimulated with LPS (or RPMI as a control). Since IL-1β enhanced response is one of the major features to characterize TI, we measured the production of IL-1β and calculated the FC between T3m and T0, stimulated by S. aureus after 24 h. Thirty-nine individuals were selected with diverse responses to TI and divided into high (FC ≥ 2; 19 individuals) and low (FC < 2; 20 individuals) responders. See also Figure S1A.
(B) UMAP visualization of cell types in PBMCs based on known marker genes and reference database. Among them, eight major cell types were identified: monocytes, CD4+ T cells, CD8+ T cells, natural killer (NK) cells, B cells, platelets, plasmacytoid dendritic cells (pDC), and myeloid dendritic cells (mDC). See also Figures S1B–S1D.
(C) Expression level of markers identified in main PBMC types.
(D) Cell proportion comparison between T0 and T3m in each cell type. Analysis was conducted using all 39 individuals, high responders, and low responders, respectively.
(E) The number of up- and downregulated TI response genes (TIGs) using all 39 individuals(left) and the same analysis were followed by dividing the individuals into high and low responders (right). Red bars represent the number of genes upregulated after being trained and blue bars represent the number of genes downregulated. Bars with a yellow icon below the x axis correspond to high responders, and bars with a dark-blue icon correspond to low responders. See also Figures S2A and S2B.