Figure 1.
Biophysical characterization of LASV GPCs derived from diverse lineages and scaffolded on I53-50A
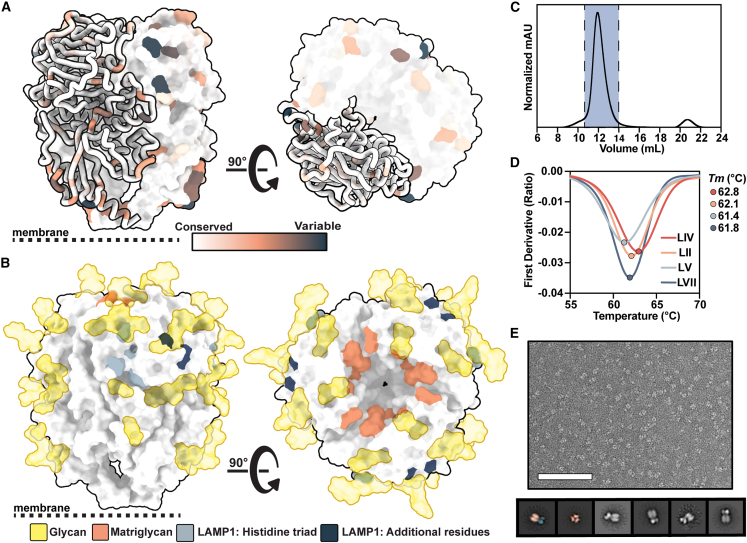
(A) LASV GPC sequence conservation mapped on ribbon and surface representation of the LIV GPC (PDB: 8EJD). Residues with increasing sequence variability are depicted in orange and dark blue.
(B) Glycans modeled from experimental density (gold; PDB: 8EJD), residues involved in matriglycan binding (orange),24 and residues suspected in LAMP-1 binding (histidine triad in gray, additional residues in navy blue40,41) mapped on the surface representation of the LIV GPC.
(C) Representative size-exclusion chromatogram (SEC) of GPC-I53-50A. Fractions containing GPC-I53-50A trimer are shown in blue.
(D) Thermostability of GPC-I53-50As assessed by the inflection point of the ratio of signal at 350 and 330 nm, as measured by nanoDSF. Circles mark the midpoint of thermal denaturation, or melting temperature (Tm), of each protein, with values listed on the right of the graph. Each melting curve is a representative of triplicate curves with Tm within ±0.1°C.
(E) Raw negative stain EM image (top) of the SEC-purified LIV GPC-I53-50A. Scale bar represents 200 nm. 2D class averages (bottom) of the GPC-I53-50A are shown with the left two classes pseudocolored to represent the GPC (orange) and I53-50A scaffold (blue).