Figure 2.
Site-specific glycosylation and structural analysis of LASV GPCs from different lineages
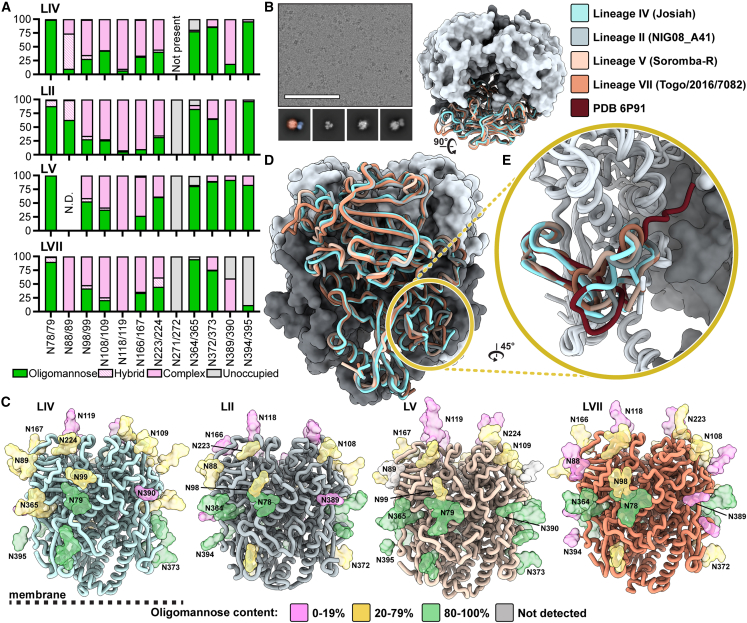
(A) Relative quantification of distinct glycan types of GPC-I53-50As determined by LC-MS describe the relative glycan processing state at a particular PNGS. Oligomannose-type glycans are shown in green, hybrid in dashed pink, and complex glycans in pink. Unoccupied sites are shown in gray.
(B) Representative micrograph of ligand-free GPC-I53-50A. Sample 2D classes are shown below, with the leftmost class pseudocolored to indicate the GPC (orange) and I53-50A trimerization scaffold (blue). Scale bar represents 100 nm.
(C) Refined atomic models of ligand-free LASV GPC structures of LIV (strain Josiah), LII (strain NIG08-A41), LV (strain Soromba-R), and LVII (strain Togo/2016/7082). Glycans are shown as colored surfaces according to their oligomannose content. Though MS data show N394 on the LVII GPC as primarily unoccupied, it is colored according to its glycan identity when present since the PNGS site was observed in the EM data. Access codes are as follows: LIV, PDB: 8EJD, EMDB: EMD-28178; LII, PDB: 8EJE, EMDB: EMD-28179; LV, PDB: 8EJF, EMDB: EMD-28180; and LVII, PDB: 8EJG, EMDB: EMD-28181.
(D) Comparison of models in (C).
(E) Comparison of fusion peptides (LIV and LV residues 260–299; LII and LVII residues 259–298) of models in (C) with PDB: 6P91,42 which features the LIV GPC in complex with 18.5C Fab.