Figure 1.
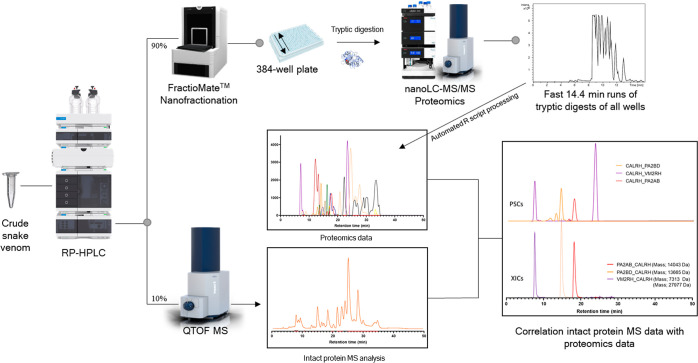
Graphical overview of the high-throughput venomics workflow. First, snake venom was subjected to nanofractionation analytics, which involves liquid chromatographic separation of venom toxins followed by a flow split of 10% to mass spectrometry (MS) for intact toxin analysis and 90% to parallel high-resolution fractionation of the separated venom toxins on to a 384-well plate. After vacuum-centrifugation of the well plate to evaporate the eluents, a tryptic digestion procedure is performed directly on the well plate using automated pipetting steps. The well plate is then directly transferred to nanoLC–MS/MS for analysis using a fast-analytical gradient runtime of 14.4 min, resulting in 100 measurements per day. The proteomics data obtained are then automatically subjected to Mascot database searching using Mascot Daemon. Next, using in-house written R scripts, all Mascot data are compiled into a single Excel sheet in which information on toxins identified is sorted by fractionation time (i.e., retention time of elution for each toxin; all toxins have eluted over a series of subsequent wells during the high-resolution fractionation). From there, for each identified toxin, a script plots so-called Protein Score Chromatograms (PSCs), in which protein scores are plotted on the y-axis versus retention times of adjacent series of wells in which a toxin was fractionated on the x-axis. The peaks in all PSCs were subsequently integrated to yield semiquantitation results on the toxins in a venom under study. Finally, the obtained venomics and intact MS data could be correlated for additional toxin characterization.