Extended Data Fig. 5 |. Design of small libraries and reproducibility of base editing efficiencies using these libraries.
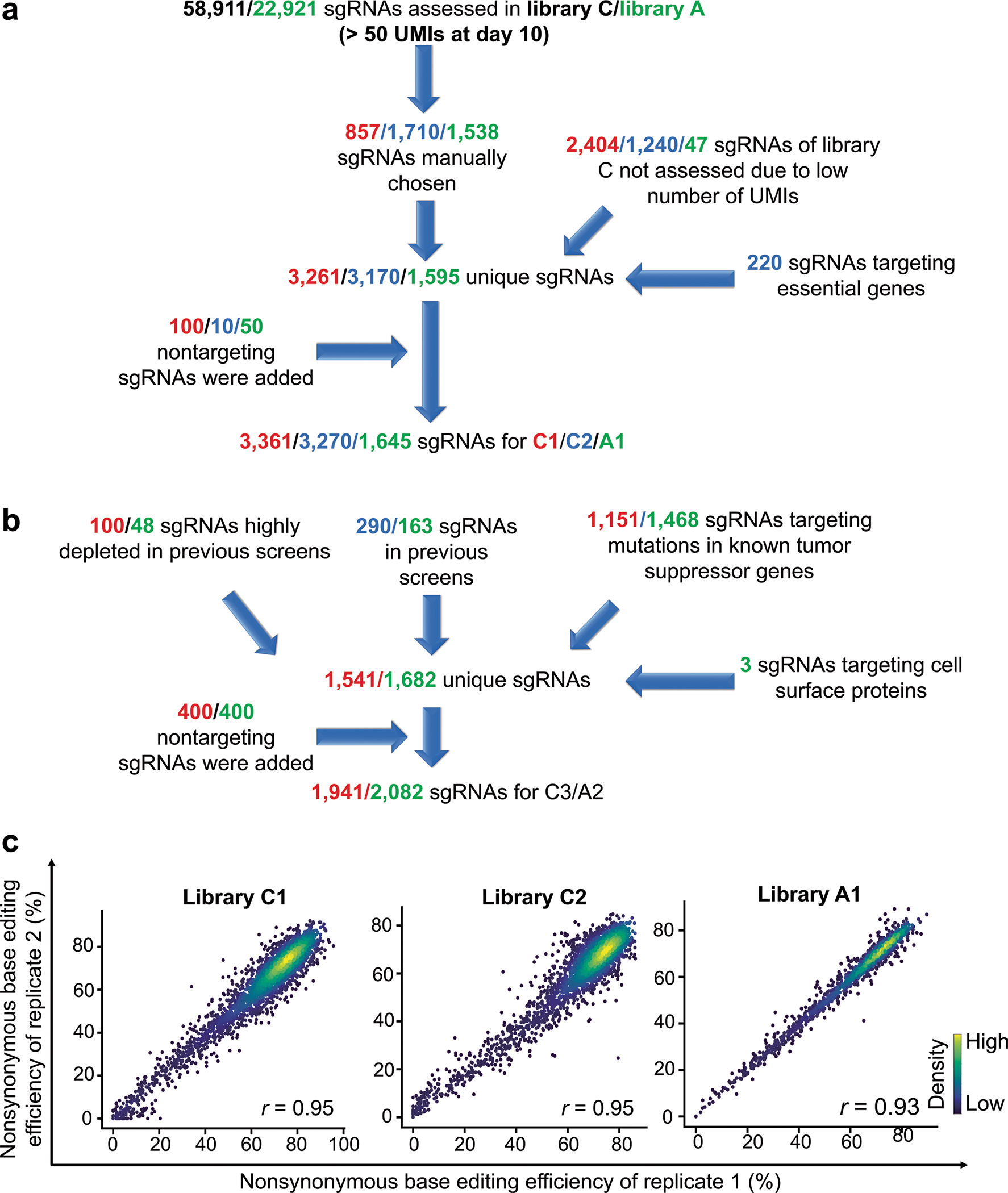
a-b, Design of small libraries C1, C2, and A1 (a) and C3 and A2 (b). UMIs, unique molecular identifiers. c, Correlations between nonsynonymous base editing efficiencies at the integrated target sequences of biological replicates. The color of each dot was determined by the number of neighboring dots (that is, dots within a distance that is three times the radius of the dot). The base editing efficiencies were determined ten days after the initial transduction of each library into P-C or P-A cells. Only sgRNAs with more than 100 raw read counts in each replicate were included. Pearson correlation coefficients (r) are shown. The number of sgRNAs n = 3,181 (library C1), 3,063 (library C2), and 1,520 (library A1).
