Fig. 2 |. Functional classification of cancer-associated transition mutations.
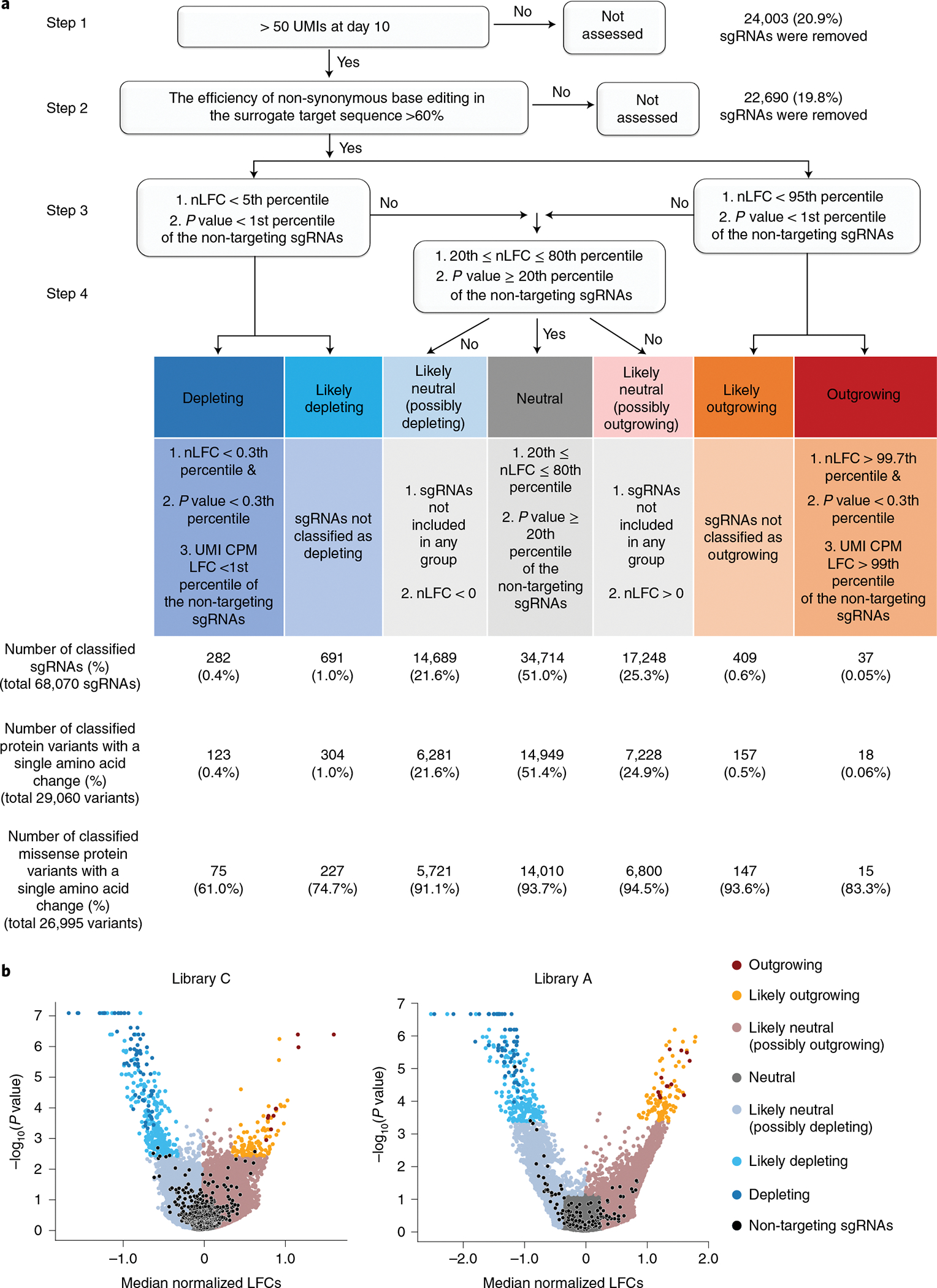
a, Functional classification process. (Step 1) sgRNAs harboring more than 50 UMIs were used as inputs for MAGeCK analysis. (Step 2) sgRNAs associated with a non-synonymous editing efficiency of less than 60% in the integrated target sequence were eliminated. (Step 3) sgRNAs were grouped depending on their nLFCs and P values obtained from MAGeCK-UMI analyses. The cutoff value was determined by the distribution of the non-targeting controls in each library. (Step 4) For the outgrowing and depleting groups, UMI CPM LFCs were further considered to prevent false classification into outgrowing and depleting groups. The number of sgRNAs and mutant proteins classified in each group are shown in the chart (integrated results based on libraries C, C1, C2, C3, A, A1, A2 and dA are shown). b, Volcano plots of nLFCs and negative logarithm of RRA P values of sgRNAs. The colors of the dots (sgRNAs) represent their functional classifications. Non-targeting sgRNAs are shown in black.
