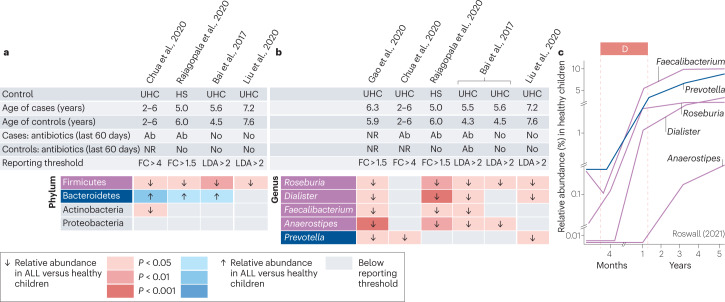
Fig. 3. Case–control studies investigating differences in gut microbiome composition at the level of phylum and genus between healthy children and children with newly diagnosed acute lymphoblastic leukaemia.
We systematically searched the MEDLINE and Embase databases from inception until 20 October 2022 to identify existing clinical studies examining the diversity and composition of the gut microbiome of children with newly diagnosed acute lymphoblastic leukaemia (ALL). The search strategy and the criteria for study selection are provided in Supplementary Table 2 and in Supplementary methods. Only for the study of Liu et al. (2020), we used publicly available participant-level data to analyse differences in relative abundance between study groups at the level of phylum and genus using linear discriminant analysis of effect size (LEfSE) as described in Supplementary methods. For all other studies, data were directly extracted as reported in individual publications (Gao et al. 2020 (ref. 56), Chua et al. 2020 (ref. 75), Rajagopala et al. 2020 (ref. 73), Bai et al. 2017 (ref. 74) and Liu et al. 2020 (ref. 70)). a, Differences in the relative abundance of different bacterial phyla between children with ALL at the time of diagnosis and healthy children. b, Selected bacterial genera showing consistent differences in their relative abundance in children with ALL compared with healthy children. Only genera with consistent differences between study groups across three or more independent studies are presented. The complete list of genera identified by all studies (including known developmental trajectories) is provided in Supplementary Fig. 2.c, Physiological changes in the relative abundance of the five identified genera in healthy children during early childhood as reported by Roswall et al.61. Ab, antibiotics; FC, fold change; HS, healthy sibling; LDA, linear discriminant analysis score; NR, not reported; UHC, unrelated healthy children.