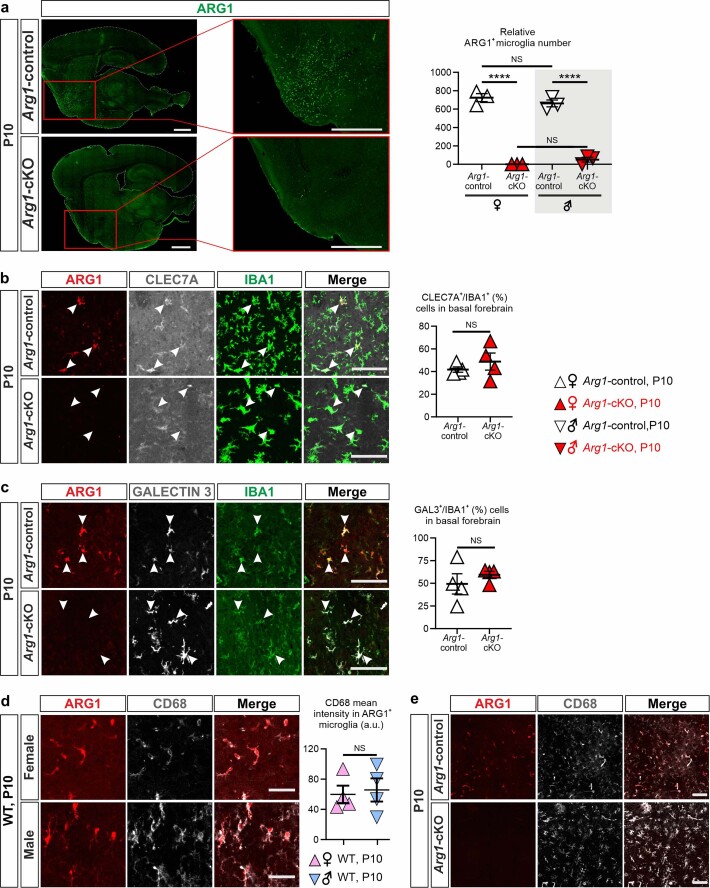
Extended Data Fig. 8. Arg1+microglia are efficiently knocked down in female and male Arg1-cKO.
a, Staining of matching sections shows that in Arg1-cKO animals, only few Arg1+microglia remain when compared to Arg1-Control. Quantification of Arg1+microglia from matching sections (n = 3 animals per group). Female, Arg1Control – Arg1cKO, P < 0.0001; male, Arg1Control – Arg1cKO, P < 0.0001. Scale bar, x = 1000 μm. b, Arg1+microglia co-localize with the marker GALECTIN-3, as inferred by our RNA-Seq analysis. Scale bar, x = 100 μm, z = 10 μm. In Arg1-cKO basal forebrain, the percentage of CLEC7A +/IBA1 + cells is not statistically significant different to the percentage of CLEC7A + /IBA1 + cells in the basal forebrain of Arg1-Control animals. c, Similar for GALECTIN-3 + /IBA1 + cells. d, Mean intensity of the lysosomal marker CD68 in female and male Arg1+microglia in BF. The images in b, c and e are representative of 4 female animals per group. Scale bars, x = 50 μm, z = 5 μm. e, CD68 expression in BF of Arg1-Control and Arg1-cKO basal forebrain. Scale bars, x = 100 μm, z = 5 μm. Data in a-d are represented as mean ± s.e.m. Statistically significant differences were determined by ANOVA (a), Mann-Whitney U (b), or unpaired t tests (c and d). All P-values are two-sided, and P-values for multiple comparisons were corrected using Bonferroni’s method (a). ****P < 0.0001, n.s. indicates not significant.