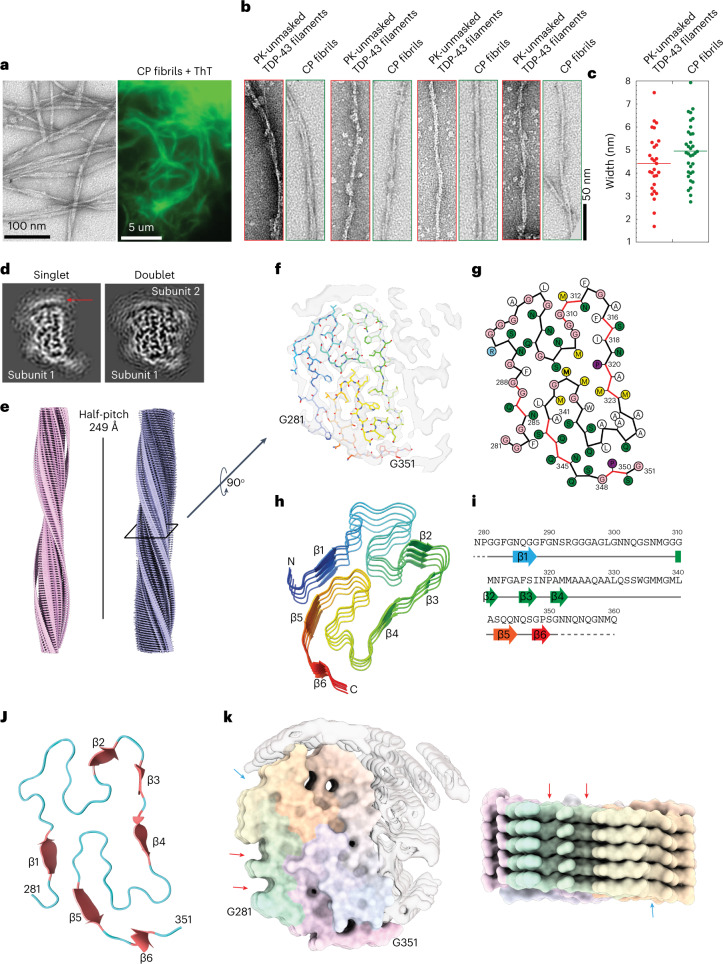
Fig. 5. Morphological properties and cryo-electron microscopy structure of the core peptide 279–360 fibrils.
a, EM image and wide-angle fluorescence microscopic image of ThT-positive CP fibrils. Images represent the data from at least three independent repeats. b,c, EM images showing morphological similarities between PK-unmasked TDP-43 filaments and CP fibrils (b) and width distribution (c). d, Central slice of 3D reconstructions of two major populations (singlet and doublet) of CP fibrils. Disconnected density from the second protofilament on top of the singlet is highlighted with a red arrow. e, Cryo-EM 3D reconstructions of singlet and doublet CP fibrils with a half-pitch of 249 Å. f, Central slice of 3D reconstruction (transparent, EMDB-13795) with one layer of an atomic model (G281-G351, PDB 7Q3U). g, Scheme of one cross-sectional layer of the CP atomic model. h, Tilted view of five neighboring subunits of CP filament. Six β-strands of the top subunit are shown. i, Schematic of the sequence and secondary structure of the CP fibrils. Six β-strands are represented by arrows colored from blue to red, loops/turns represented by a gray line, and the unsolved residues as the dashed lines. j, One cross-sectional layer of the atomic model revealing the ‘H-fold’ conformation of the CP fibrils. Six β-strands are shown as thicker red arrows with the loops in blue. k, Surface representation of CP fibrils with the arrows pointing to the shallow grooves on the sides of the CP fibril structure.