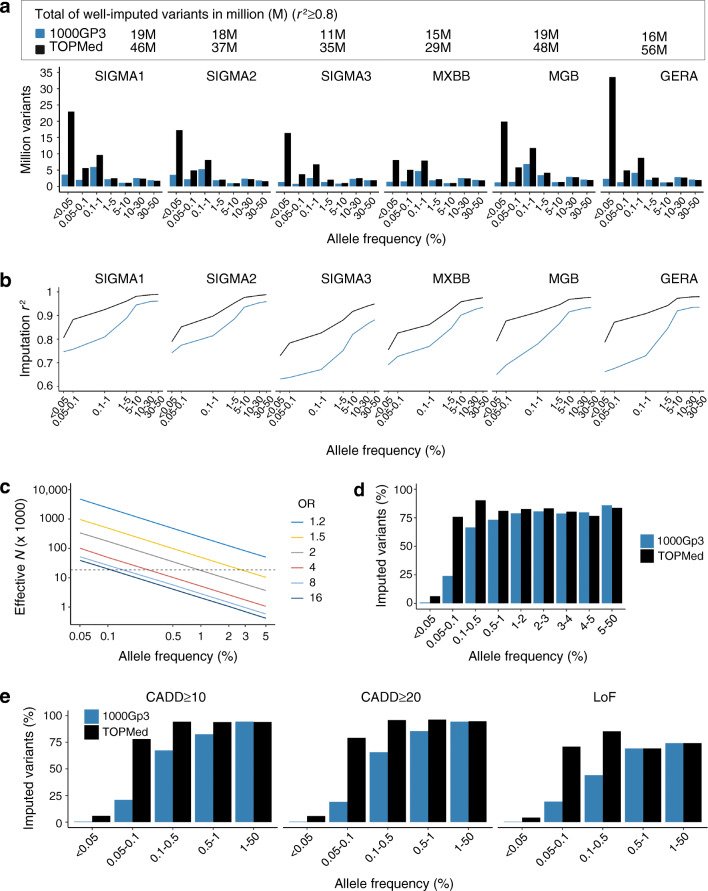
Fig. 2.
Performance of the TOPMed reference panel for the imputation of Latino samples. (a) Number of chromosome-wide well-imputed variants (imputation r2≥0.8) by AF for each analysed cohort when using the 1000GP3 (blue) or the TOPMed (black) reference panels. (b) Average chromosome-wide imputation quality by AF for each analysed cohort when using the 1000GP3 (blue) or the TOPMed (black) reference panels. (c) Effective sample size required for reaching 80% statistical power to detect genome-wide significant signals at different effect sizes (OR). The dotted lines show the discovery effective sample size of this study. (d) Percentage of the exome sequenced variants in chromosome 22 that could be imputed when using the 1000GP3 (blue) or the TOPMed (black) reference panels. (e) Percentage of the exome sequenced LoF and deleterious predicted variants based on CADD score in chromosome 22 that could be imputed when using the 1000GP3 (blue) or the TOPMed (black) reference panels