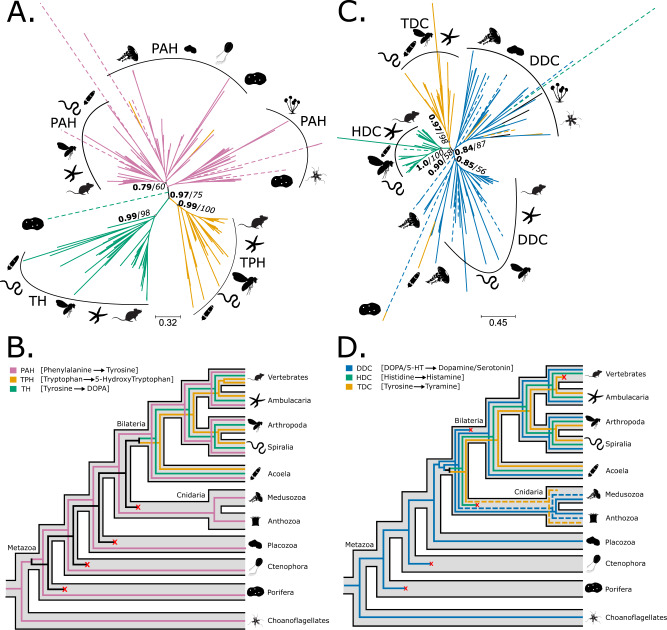
Fig. 2. Phylogeny and reconciliation for aromatic amino acid hydroxylases (AAAHs) and amino acid decarboxylases (AADCs).
A Transfer bootstrap expectation tree and (B) simplified illustration of reconciliation calculated using Generax for AAAH sequences. C Transfer bootstrap expectation tree and (D) simplified illustration of reconciliation calculated using Generax for AADC sequences. The nodal supports shown are transfer bootstrap expectation (TBE) scores (in bold), and ultrafast bootstrap proportion supports (in italic) for key nodes. Dashed lines indicate sequences identified as unstable in the t-index and leaf stability index (LSI) analysis (see Supplementary Data 6 for details). PAH phenylalanine hydroxylase, TPH tryptophan hydroxylase, TH tyrosine hydroxylase, DDC dopa decarboxylase, TDC tyrosine decarboxylase, HDC histidine decarboxylase. Silhouettes obtained from Phylopic.org. Silhouette images are by Andrew R. Gehrke (Hofstenia miamia); Daniel Jaron (Mus musculus); Mali’o Kodis, photograph by Ching (http://www.flickr.com/photos/36302473@N03/) (Chrysaora fuscescens); Mario Quevedo (Asteriidae); Oliver Voigt (Trichoplax adhaerens); Ramiro Morales-Hojas (Drosophila americana); Steven Haddock, Jellywatch.org (Hormiphora californensis); and Tess Linden (Salpingoeca rosetta). Source Data are available at [https://figshare.le.ac.uk/articles/dataset/Monoamine_Neuromodulation_is_a_Bilaterian_Innovation_Results/20391477] in Results/GeneTrees labelled OG_AAAH_TBE.tbe.tree, OG_AAAH_UFB.treefile, OG_AADC_TBE.tbe.tree and OG_AADC_UFB.treefile.