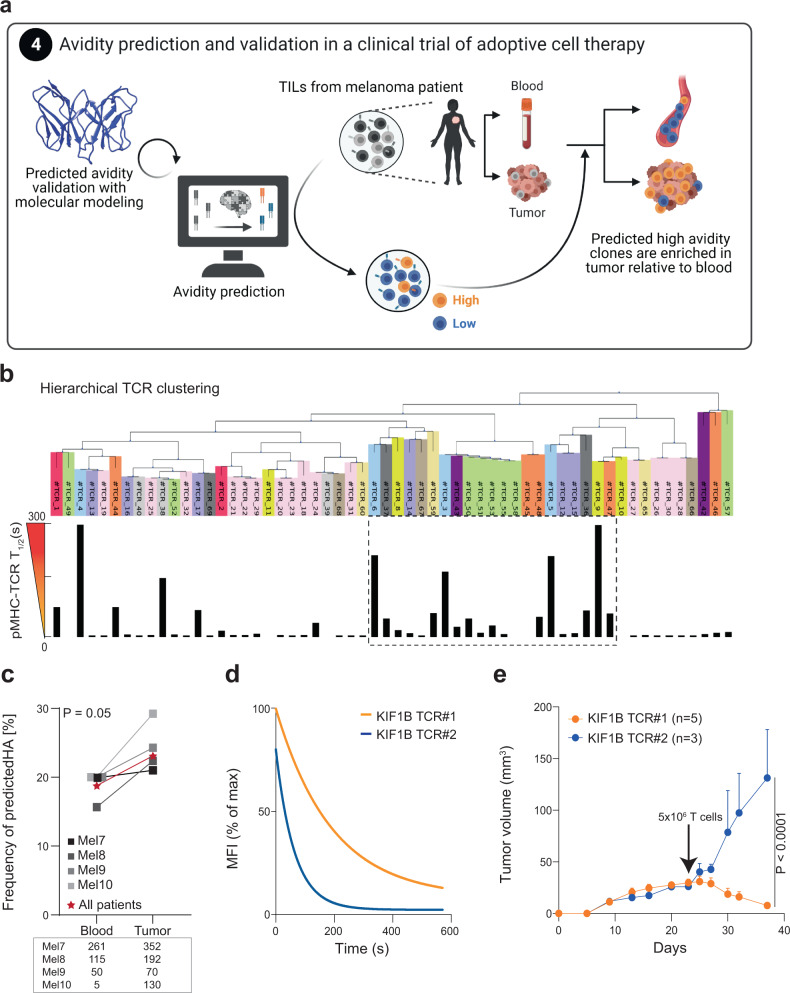
Fig. 4. Tumor infiltration after ACT correlates with predicted structural avidity inferred from TCR clustering analyses.
a Computational analysis of TCR features led to the establishment of a predictor of TCR avidity and its application on patients’ TIL-ACT products allowed tracking of predicted low and high-avidity TCRs in post-ACT tumor samples. b Hierarchical clustering of 58 TCR sequences provided based on a biophysical approach43. TCRs sharing the closest 4-mer features are next to each other and TCRs recognizing the same pMHC have the same color code. TCR model numbers are presented as labels and further details about TRAV, TRAJ, TRBV, TRBJ, HLA and peptide are found in Supplementary Table 4. The structural avidity of each cognate TCRs is represented below (mean of n = 3 independent experiments). The black dashed box highlights a region where high-avidity TCRs recognizing multiple pMHC specificities are clustering. c Cumulative analysis for four melanoma patients of the percentage of predicted high-avidity CD8 T cells in blood and tumor samples. Values for individual patients are plotted (gray and black) as well as the cumulative analysis (in red) for which the P value was calculated as described in the method section. The number of clones is indicated below for each patient individually. d Monomeric pMHC-TCR dissociation kinetics of Jurkat cells transfected with neoantigen KIF1BS918F-specific TCR#1 and TCR#2, respectively predicted as high and low-avidity TCRs. e Autologous (Mel8) tumor growth in IL-2 NOG mice adoptively transferred at day 22 with 5 × 106 primary T cells transduced with KIF1BS918F-specific TCRs (n = 1 independent experiment, Mean ± SEM). Log-rank two-sided test was used to determine the exact P value. Source data are provided as a Source Data file.