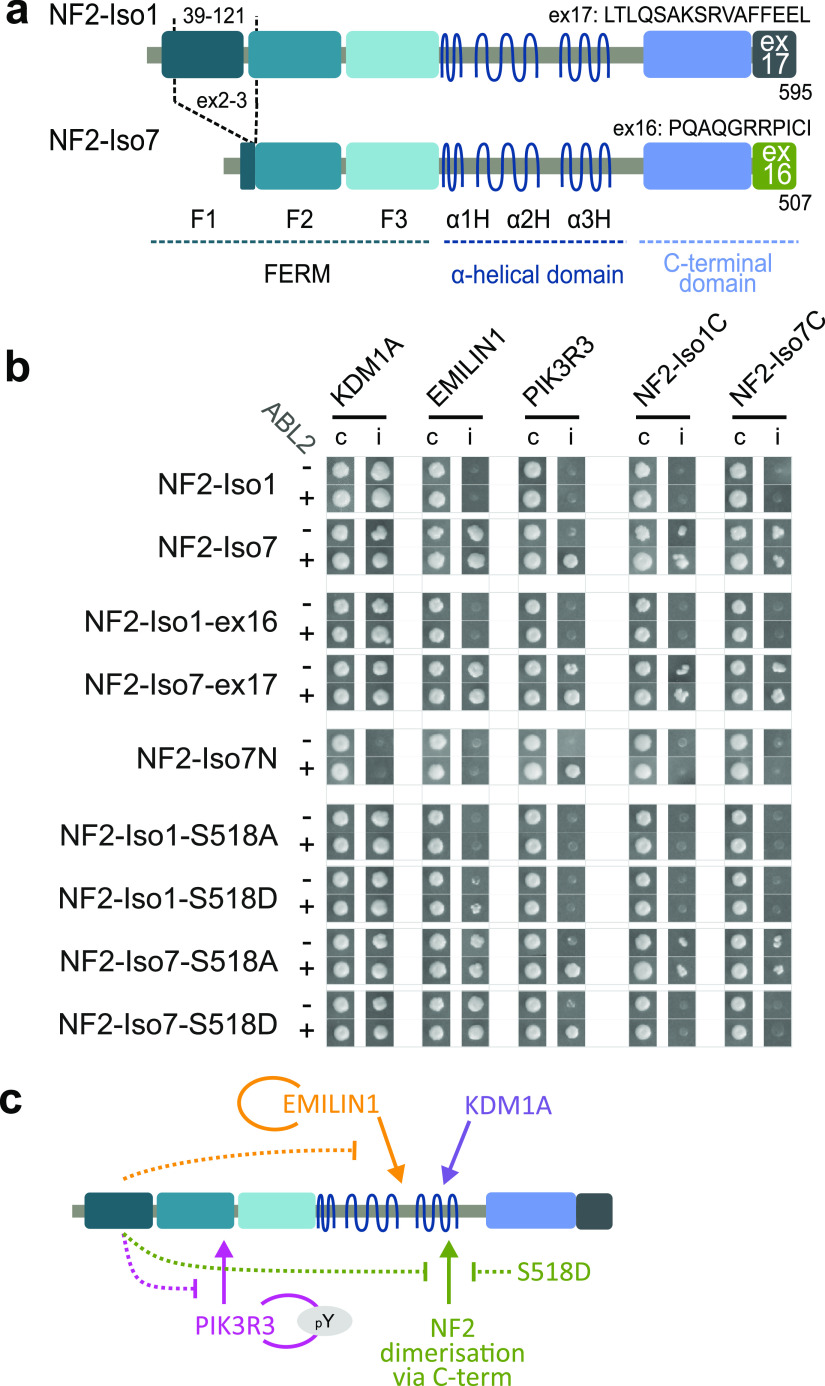
Figure 1. Conformation dependent PPIs of NF2.
(A) NF2 splice isoforms. The canonical isoform 1 is 595 amino acids long, the C-terminus is derived from sequences of exon 17 (595 AA [ex17], P35240-1). NF2 isoform 7 is a shorter splice isoform (507 AA [Δex2/3, ex16], P35240-4). The C-terminus is alternatively spliced and includes exon 16-derived sequences which are 5 amino acids shorter (iso-1 580–590: LTLQSAKSRVA → PQAQGRRPICI, iso-1 590–595 missing), isoform 7 lacks most of the F1 FERM subdomain and the beginning of the F2 subdomain encoded in exons 2 and 3, amino acids 39–121. Tumor suppressor activity has been demonstrated for isoform 1 but not for isoform 7. (B) Y2H protein interaction results. Indicated NF2 bait constructs (row, see Fig S1A) were tested with five different prey constructs (columns) in the presence and absence of active tyrosine kinase ABL2 (+) and ABL2KD (K35M) (−) respectively. Note: NF2-Iso1N (1–332) was autoactive and was excluded from the analysis. Prey: KDM1A (O60341), EMILIN1 (Q9Y6C2), PIK3R3 (Q92569), NF2-Iso1C (P35240-1: 308–595), NF2-Iso7C (P35240-4: 225–507). Growth of diploid yeast on non-selective agar (c) and on selective agar indicating protein interactions (i) is shown. (C) Graphical summary of the interactions observed. Interactions with NF2 are shown with arrows; conformational interaction inhibition is indicated with dashed lines.