Figure 4. Structural analyses of NF2 variants.
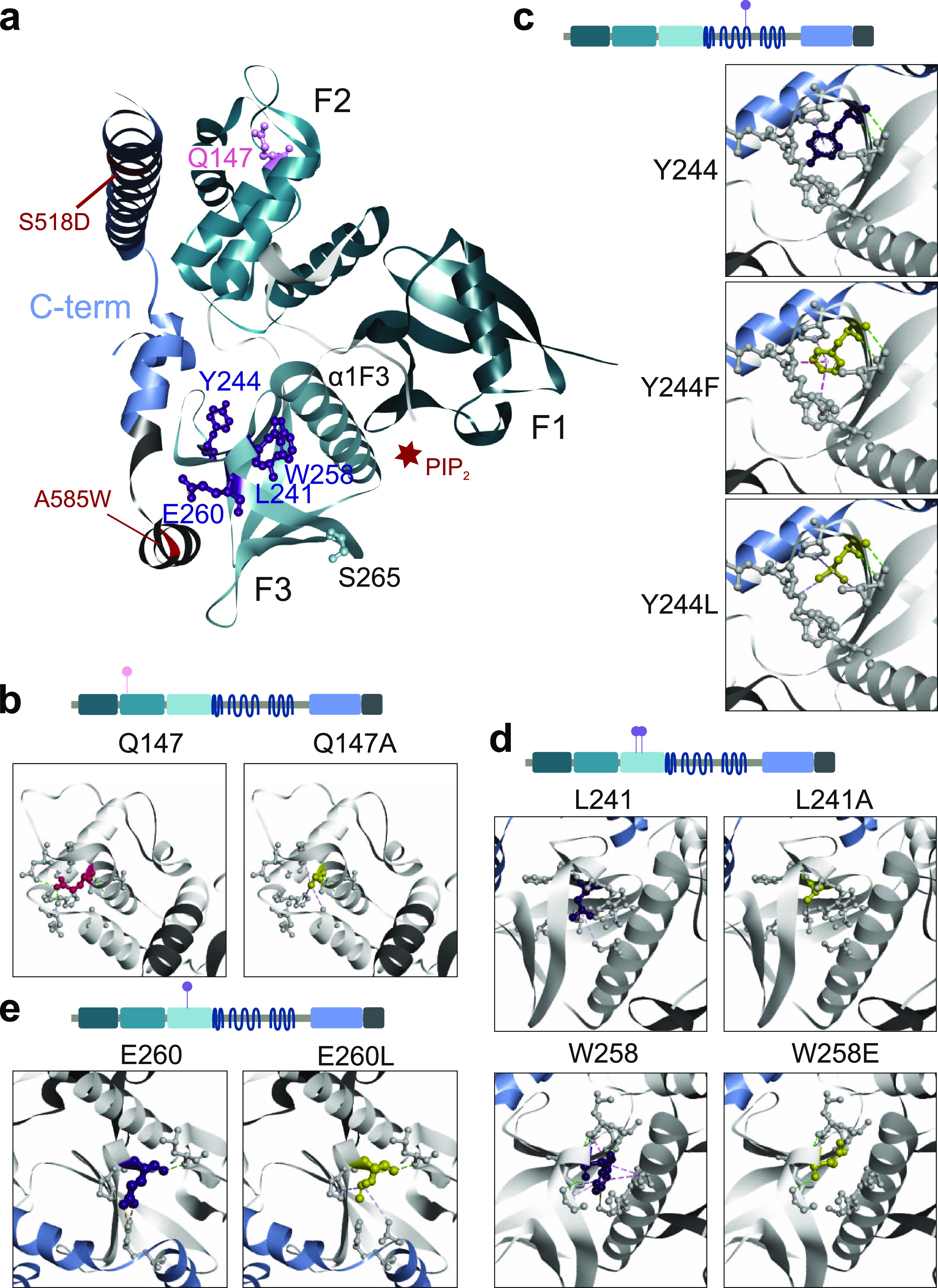
(A) FERM domain mutations projected on the NF2 protein structure. 3D ribbon “cloverleaf” structure of the FERM domain in grey, C-terminal NF2 peptide in blue colors (PDB: 4zrj). Mutations S518D and A585W in the C-terminal NF2 peptide that stabilized the structure are indicated, and the PIP2 binding groove between F3 and F1 subdomains (red star). Amino acid positions perturbing the NF2 interaction pattern (purple, ball and stick) cluster in the F3-FERM subdomain. (B, C, D) 3D structures of the FERM domain as ribbon models in which indicated residues are colored and displayed as ball and stick atomic models with the respective interactions indicated as dashed lines. WT residue (left panel) in comparison with mutant residue (right panel). (B): Q147; (C): Y244, (D): L241 and W258, (E): E260. The models were created with BIOVIA Discovery Studio 2020 (version 20.1.0.19295).
