Abstract
Despite the promise of combination cancer therapy, it remains challenging to develop targeted strategies that are nontoxic to normal cells. Here we report a combination therapeutic strategy based on engineered DNAzyme molecular machines that can promote cancer apoptosis via dynamic inter- and intracellular regulation. To achieve external regulation of T-cell/cancer cell interactions, we designed a DNAzyme-based molecular machine with an aptamer and an i-motif, as the MUC-1-selective aptamer allows the specific recognition of cancer cells, and the i-motif is folded under the tumor acidic microenvironment, shortening the intercellular distance. As a result, T-cells are released by metal ion activated DNAzyme cleavage. To achieve internal regulation of mitochondria, we delivered another DNAzyme-based molecular machine with mitochondria-targeted peptides into cancer cells to induce mitochondria aggregation. Our strategy achieved an enhanced killing effect in zinc deficient cancer cells.
Keywords: DNAzyme, Molecular machine, Dynamic cellular regulation, Combination cancer therapy, Cell apoptosis
Entry for the Table of Contents

A combination therapeutic strategy based on engineered DNAzyme molecular machines was designed for promoting cancer apoptosis via dynamic inter- and intracellular regulation. Our strategy resulted in an enhanced killing effect, demonstrated using zinc deficient cancer cells, thus providing a promising next-generation combination therapy for cancer.
Introduction
Cancer is a major cause of death in the world and numerous efforts have been devoted to the development of better cancer treatment methods.[1] To improve the treatment effect, a typical strategy is to use high dosages of nonspecific chemotherapy drugs such as doxorubicin, paclitaxel, and methotrexate. However, extensive usage of such drugs may cause severe side effects by causing damages to normal cells.[2] To reduce side effects while maintaining cancer treatment efficacy, combination therapies hold great promise.[3] A typical combination therapeutic strategy is to combine immunotherapy with nanoparticle-based drug delivery, which can induce immunological killing of cancer cells with increased intracellular drug concentration at the same time.[4] However, the treatment outcomes of some combination therapeutic methods still fail to reach the medical frontline efficacy, especially for methods based on T-cells,[5] owing to limitations in the design of targeting agents that can specifically link between T-cells and cancer cells, as the targeting agents usually adopts large natural or artificial structures, thus may hinder the drug delivery and affect the immune response.[6]
In addition, most previous studies can only control T-cell and cancer cell recognition, even though tunable departure of T-cells from treated cancer cells is just as essential for cancer immune therapy.[7] Therefore, despite progress made in this field, it remains challenging to design an effective strategy to achieve the dynamic regulation of cell-cell interactions between T-cells and cancer cells, including cell recognition, cell-cell approaching and post-treatment separation. In addition, for intracellular treatment, it is important to reduce the side effects of drugs to normal cells. As it is well documented that the increased production of reactive oxygen species (ROS) enables anti-cancer effects, some nearest studies found that mitochondrial aggregation can generate excessive ROS, resulting in cancer cell death.[8] Although these methods have been shown to improve cancer apoptosis, their indiscriminate attack on normal cells has remained unsolved.
To overcome this limitation, we report herein a combination therapeutic strategy based on molecular machines incorporating engineered DNAzymes and DNA aptamers to induce cancer apoptosis from both the extracellular control of T-cell/cancer cell interactions and intracellular control of mitochondrial aggregation. On one hand, cell-cell interactions between T-cells and cancer cells are especially attractive to researchers, as the specific recognitions of T-cells to the cancer cells are extremely important to build an effective immune therapy.[9] On the other hand, the dispersion of mitochondria in cancer cells increases cancer invasiveness,[10] while the aggregation of mitochondria is favorable to cause cancer cell death by overproducing ROS.[11] DNAzymes have the capability of catalyzing the breakage of ribonucleotides in the presence of specific metal ions.[12] Due to the abnormal level of metal ions in some highly aggressive cancers, such as zinc deficient pancreatic cancer cells, the internal regulation based on DNAzyme may aid the discovery of innovative cancer therapies with higher targeting ability and fewer side effects.[13] Taking advantages of the susceptibility of DNAzymes to metal-dependent cleavage, we have previously employed metal ion-specific RNA-cleaving DNAzymes and their respective substrate strands as building blocks to design different control switches for regulating the cell behaviors including cell-cell conjunctions and disaggregation of both individual cells and multicellular spheroids.[14] However, the cholesterol anchors and pH-sensitive fluorescence markers limited its applications due to the lack of targeting ability and quenching of fluorescence under tumor microenvironment. To overcome these limitations, we construct molecular machines by using RNA-cleaving DNAzymes modified with different functional tags, thus allowing specific interactions between cells or mitochondria to obtain a more efficient cancer combination therapy.
Results and Discussion
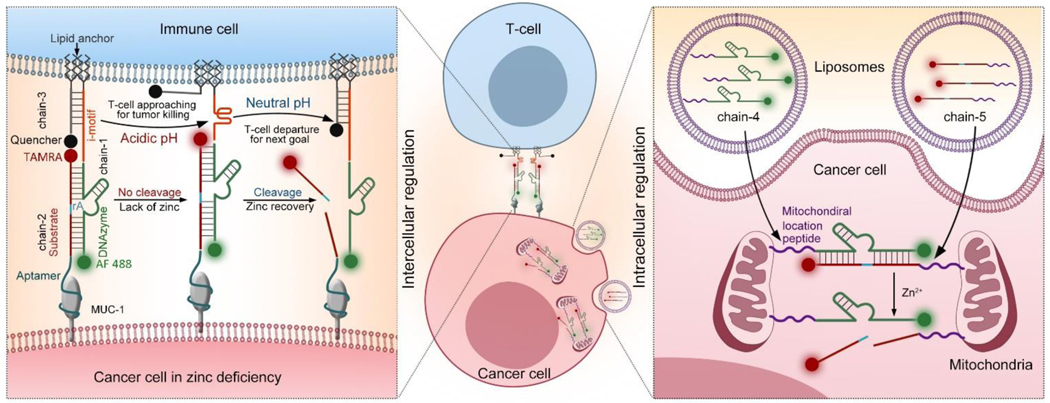
As shown in Scheme 1, for the external regulation of cell-cell interactions between T-cells and cancer cells, a Zn2+-specific DNAzyme[15] (chain-1) is modified with double lipid tails at the 5´-end, allowing the substrate chain to be spontaneously inserted into a live cell surface through hydrophobic interactions.[16] In addition, an i-motif sequence is inserted between the lipid tail and the DNAzyme part, allowing pH-sensitive structure transformation to regulate the distance between linked cells.[17] The corresponding substrate (chain-2) is modified with an aptamer at the 5´-end for the specific linkage of MUC-1 proteins overexpressed on cancer cell surface.[18] The 3´-ends of chain-1 and chain-2 are tagged with fluorescent markers (Alexa Fluor 488 (AF 488) for chain-1 and carboxy tetramethyl rhodamine (TAMRA for chain-2). AF 488 and TAMRA are stable at acidic pH, which is favorable for real-time fluorescence tracking under tumor acidic microenvironment.[19] To monitor the distance between T-cells and cancer cells, a complementary strand of i-motif with a lipid anchor at 3´-end (chain-3) is attached to the T-cell surface and hybridized with the i-motif part of chain-1. A corresponding quencher of TAMRA, BHQ2, is tagged at the 5´-end of chain-3. The cell-cell distance can be revealed by the efficiency of the quenching effect between TAMRA and BHQ2.[20] Through the hybridization of these three DNA chains, a DNAzyme molecular machine allowing precise regulation of cell-cell interactions between T-cells and cancer cells is obtained, enabling cancer cell recognition, T-cell/cancer cell approaching, and post-treatment separation. Engineered with aptamer, i-motif, and FRET pairs, the DNAzyme molecular machine is capable to sense and respond to the tumor microenvironment.[21] Using the tumor marker MUC-1 as the target protein,[22] the aptamer sequence allows the specific recognition of cancer cells.[23] Next, the tumor acidic microenvironment triggers the transformation of i-motif sequence from a straight strand to a folded structure, shortening the distance between cancer cells and T-cells to induce the killing of cancer cells. Then T-cells are released by metal ion (Zn2+) activated DNAzyme cleavage, enabling quick release of T-cells from treated cancer cells. During the whole process of external regulation, the cell-cell distance can be revealed by monitoring the fluorescence intensity of TAMRA. Under a weak alkaline pH value, the i-motif sequence kept straight, thus the TAMRA is closer to and quenched by its quencher. When the pH value is acidic, the i-motif folds, which cause the releasing of TAMRA from the quencher and light up.
Scheme 1.
Schematic illustration showing combination cancer treatment based on engineered DNAzyme molecular machines. (Left) Dynamic intercellular regulation of T-cell/cancer cell interactions based on chain-1, chain-2 and chain-3. The distance between T-cells and cancer cells can be revealed by the fluorescence signal (TAMRA). (Right) Dynamic intracellular regulation of mitochondria aggregation in cancer cells based on chain-4 and chain-5. Here, the mitochondria aggregation only happened in zinc deficient cancer cells.
To further realize the internal regulation of intracellular mitochondria, the enzyme and substrate strands of Zn2+-specific DNAzyme are wrapped into liposomes separately, and both DNAzyme (chain-4) and its substrate (chain-5) are modified with a mitochondrial localization peptide tail at the 5´-end, allowing mitochondrial targeting.[24] Via the hybridization of chain-4 and chain-5, the separated mitochondria start to link with each other and form large mitochondria aggregates, enabling overproduction of intracellular ROS to promote cancer cell death. The disassembly of mitochondria can be controlled by Zn2+ ions mediated substrate cleavage. Thus, a DNAzyme molecular machine allowing dynamic control of mitochondria behaviors is achieved. Compared with a zinc deficient environment in cancer cells, which can reduce the cleavage of substrate, thus favors the hybridization of these two chains, DNAzyme cleavages happens in normal cells having normal zinc levels. The melting temperatures of the DNAzymes are approximately 70 °C, much higher than 37 °C where the cells are cultured; thus, the substrates remain tightly bound. The halves of the cleaved substrates have much lower melting temperatures of approximately 20 °C; therefore, the substrates will be dissociated at 37 °C upon metal treatment (Table S1). The characterization of these sequences were provided in Figure S1–2. As a result, this design is favorable to reduce cytotoxicity in non-target cells to avoid side effects.[25]
Using pancreatic cancer cells (PANC-1), which is zinc deficient endogenously, as proof-of-concept,[26] we aimed to achieve the dynamic control of T-cell/cancer cell interactions and intracellular mitochondria interactions by the engineered DNAzyme molecular machines. Taken together, this strategy aims to achieve tumor microenvironment-responsive dynamic control of cell behaviors from both outside and inside, thus paving the way towards the further development of combination cancer therapy.
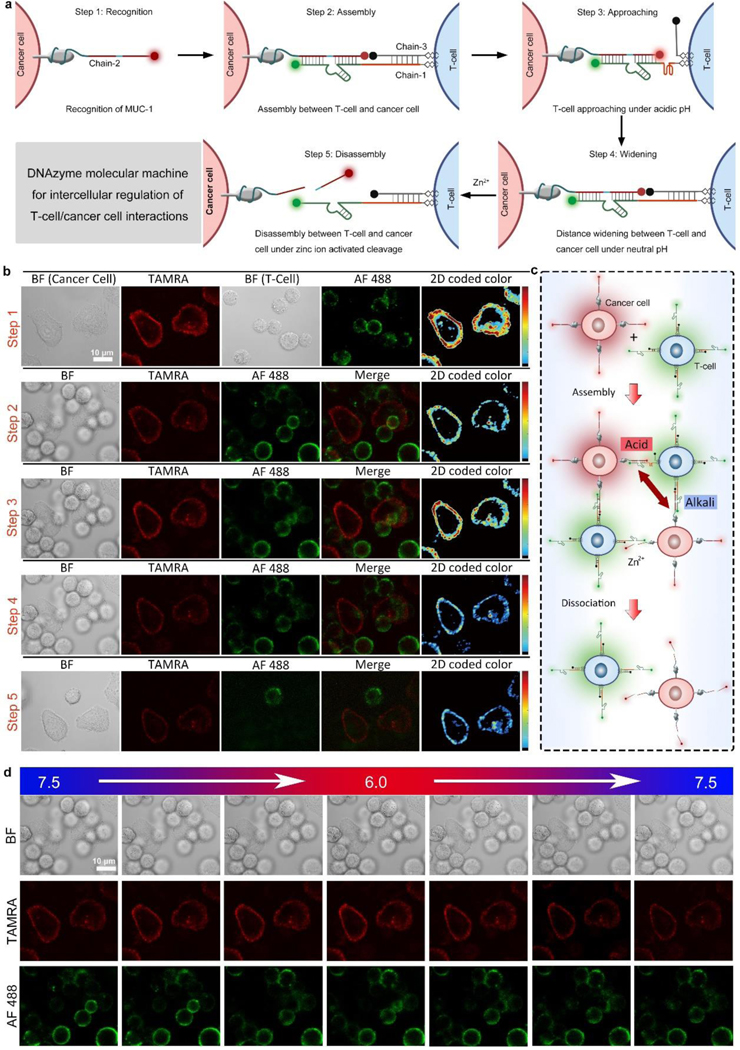
Before cell experiments, first we validated the pH-sensitive structure change of i-motif and DNAzyme-catalyzed substrate cleavage by gel electrophoresis. The mixture of chain-1 (containing a sequence of Zn2+-specific DNAzyme and i-motif), chain-2 (containing fragments of substrate and MUC-1 aptamer), and chain-3 (containing a complementary strand of i-motif) were treated with H+ or Zn2+, respectively. Both H+ and Zn2+ treatment led to the emergence of a band with lower molecular weight, indicating the departure of chain-3 under acidic pH and the cleavage of chain-2 by Zn2+ (Figure S3). For the following cell-cell interaction tests, T-cells modified with chain-1/chain-3 and cancer cells modified with chain-2 were mixed and T-cell/cancer cell assembly could be achieved via the hybridization between chain-1, chain-2, and chain-3. As shown in Figure 1a, the DNAzyme molecular machines constructed by chain-1, chain-2 and chain-3 were applied for the external regulation of T-cell/cancer cell interactions by the following five steps: recognition, assembly, approaching, widening, and disassembly. The dynamic regulation of cell-cell interactions between T-cells and cancer cells corresponding to the above five steps was confirmed by cell microscopic images (Figure. 1b). When the cells were mixed, cell-cell assembly could be achieved instantly (Video S1). After different incubation times (0 min-60 min), the cells were sent for flow cytometry analysis (Figure S4), and the fluorescence intensity reached the maximum after 30 min incubation, which was in accordance with the confocal fluorescence images (Figure 1b, step 1). After the cell-cell assembly, the fluorescence of TAMRA was quenched by BHQ2 (Figure 1b, step 2), indicating a proper insertion and hybridization of three chains. Only a weak TAMRA signal could be detected by flow cytometry analysis, which was corresponding well with the confocal fluorescence images (Figure S5a, Figure 1b, step 2). As we decreased the solution pH to 6.0, the TAMRA fluorescence was obviously restored as the acidic pH induced the folding of chain-1, causing the departure of BHQ2 at the 5´-end of chain-3 (Figure 1b, step 3).The same trends were detected by flow cytometry analysis (Figure S5b). After the pH was tuned back to neutral, the TAMRA fluorescence was quenched again due to the structure recovery of chain-1 (Figure 1b, step 4). Next, with the addition of Zn2+, substrate chain-2 was cleaved from the ribonucleotide cleavage site, causing immediate cell disassembly (Video S2), as T-cells were cut from cancer cells (Figure 1b, step 5). The dynamic regulation of cell-cell interactions between T-cells and cancer cells could be clearly shown from the histogram of the average red and green fluorescence of T-cells and cancer cells (Figure S6). The loading amounts of dye-tagged DNA strands on the cell surface were optimized (Figure S7), and the loading amount of chain-1 and chain-2 on the surface of cancer cell was calculated to be 4.96×109 and 5.51×109 molecules per cell respectively, while the loading amount of chain-2 on the surface of PANC-1 cell was 4.98×109 molecules per cell. (Figure S8, Table S2). The stability of the fluorescent markers tagged on DNA strands were tested at different pH values,[27] and the results confirmed the good stability of AF 488 and TAMRA tagged on DNA strands (Figure S9). A control experiment using chain-2 without MUC-1 aptamer confirmed the targeting ability of the aptamer part, as the cell surface fluorescence was significantly reduced (Figure S10). Next, we evaluated the targeting specificity of chain-2 to PANC-1 cells. PANC-1 cells and HEK293 cells were treated with chain-2 (tagged with TAMRA), and then tested by confocal microscopy and flow cytometry. As shown in Figure S11, the average fluorescence of PANC-1 cells was much higher than normal cells, which confirmed the targeting ability of chain-2 to PANC-1 cells. Another control experiment using chain-2 with mutated MUC-1 aptamer part (chain-20) was performed by flow cytometry (Figure S12), and the results showed neglectable fluorescence of chain-20 treated PANC-1 cells. To evaluate the release efficiency of T-cells, the interaction between T-cells and cancer cells without the addition of Zn2+ was tested. As shown in Figure S13, the T-cells remained to be connected with the cancer cells after 1 h. When the mutation was introduced to DNAzyme or substrate, the T-cell/cancer cell assemblies could not be disassembled, indicating the catalytic function caused cleavage by DNAzyme (Figure S14–15).[28] A positive control using RNase A showed similar disassembly suggested that the cleavage of adenine ribonucleotide (rA) in the substrate was likely to induce the cell disassembly (Figure S16). In addition, other metal ions (Na+, K+, Mg2+, Cu2+) did not induce the cell disassembly, indicating Zn2+-specific cleavage (Figure S17). Besides, the cleavage rate was obviously increased with the increased concentration of Zn2+ (Figure S18), which suggests that the T-cells can be cut from cancer cells when zinc ion level in the microenvironment increases to normal, allowing T-cells to leave killed cancer cells and to search for new targets. According to the above results, we chose 1 mM Zn2+ to achieve immediate cell disassembly within 10 min.
Figure 1.
Dynamic intercellular regulation. (a) Intercellular regulation of T-cell/cancer cell interactions by DNAzyme molecular machines constructed by chain-1, chain-2, and chain-3. Step 1–5: recognition, assembly, approaching, widening, and disassembly. (b) Cell images showing the dynamic regulation of cell-cell interactions between T-cells and cancer cells corresponding to Step 1–5 in (a). Color bar: 0 to 250 from bottom to top. (c) Distance control between T-cells and cancer cells by pH regulation. (d) Cell images showing the cell-cell distance changes under different pH values. BF: bight-field, TAMRA: red fluorescence, AF 488: green fluorescence, Merge: mixed green and red channel. 2D coded color: pseudo-color of red channel intensity.
Next, the reversible distance control between T-cells and cancer cells was evaluated by tuning the pH value (Figure 1c). As shown in Figure 1d, the fluorescence of TAMRA on cancer membrane was quenched under a weak alkaline pH value (pH=7.5). The TAMRA was gradually lighted up when the pH value was tuned to acidic (pH=6.0). After the pH value was gradually adjusted back to weak alkaline, the fluorescence intensity was decreased again. The average red and green channel intensity (Figure S19) confirmed that the distance between T-cell and cancer cell after linkage could be reversibly and precisely regulated by pH control. This reversible distance control could be done up to five cycles, and the performance remained satisfactory (Figure S20). Chain-1 containing multiple i-motif pieces (2–3 repeated segments) was also tested (chain-14, chain-16), and the fluorescence images indicated that one or two i-motif pieces in chain-1 could achieve ideal reversible distance control, while the fluorescence recovery was relatively weak by chain-1 containing three i-motif pieces (Figure S21–23). Therefore, in this work, we chose chain-1 containing a single i-motif piece for the experiments. A control test using T-cells modified with a DNA chain without i-motif sequence or with mutated i-motif sequence (Table S1) and cancer cells modified with chain-2 was performed to form cell assemblies, and the experimental results showed no change of red fluorescence, indicating no distance change (Figure S24–26). Moreover, cancer cells and T-cells without any modification could not form assembly, confirming the recognition efficiency of our strategy (Figure S27). Together, the above results confirmed the viability of the external regulation of T-cell/cancer cell interactions based on our design.
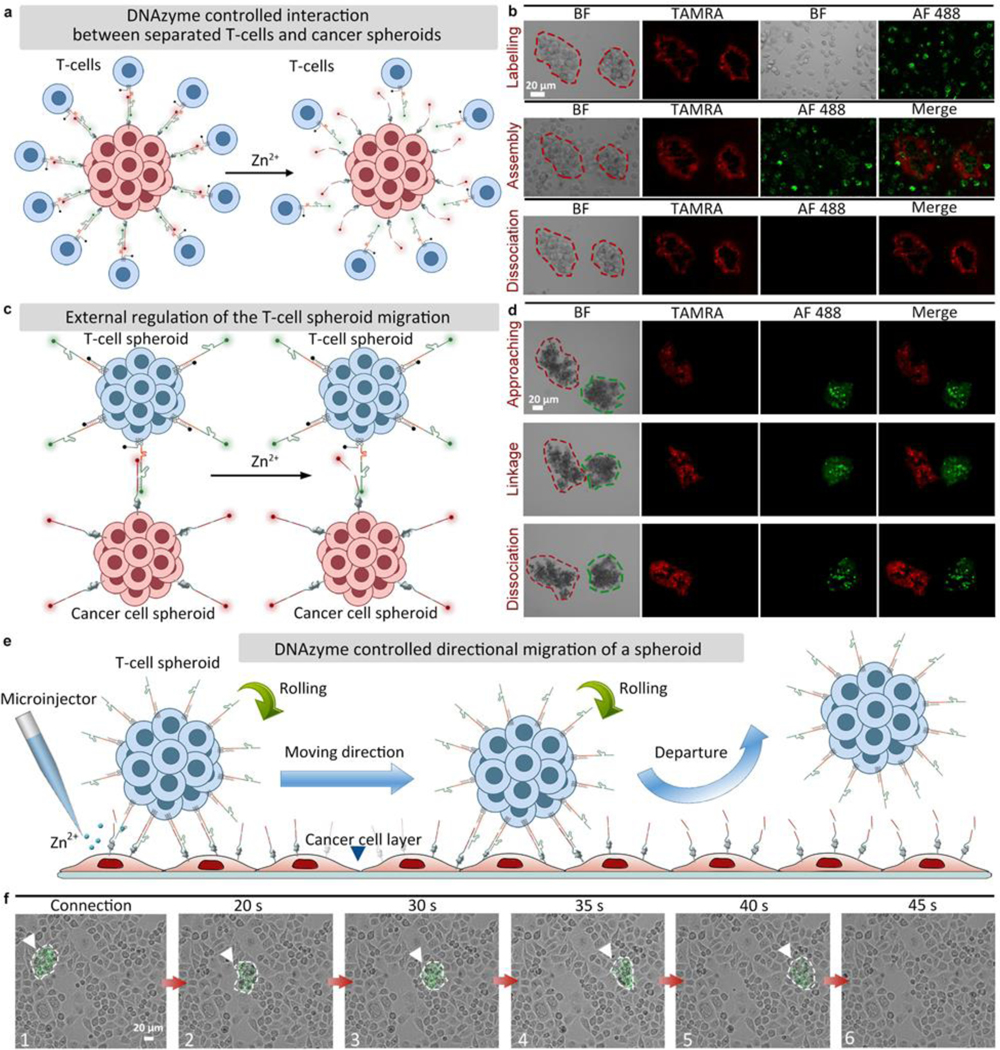
As a proof-of-concept for mimicking in vivo environments, the external regulation of T-cell/cancer cell interactions by DNAzyme molecular machines (constructed by chain-1, chain-2, and chain-3) was further investigated using cell spheroids. Firstly, DNAzyme controlled interaction between separated T-cells (chain-1/chain-3 modified) and a cancer spheroid (chain-2 modified) was realized (Figure 2a). A cancer cell spheroid modified with chain-2 could form assembly with separated T-cells modified with chain-1/chain-3. After the addition of Zn2+ ions, the T-cells could be released from the cancer spheroid (Figure 2b, Figure S28, Video S3). In contrast, cancer cell spheroid and T-cells without any modification could not form assembly (Video S4).
Figure 2.
DNAzyme controlled interaction between multicellular spheroids. (a) DNAzyme controlled interaction between separated T-cells (chain-1/chain-3 modified) and a cancer spheroid (chain-2 modified). (b) Fluorescent images showing the interactions between T-cells and a PANC-1 cancer cell spheroid (chains labeling, assembly, and dissociation under Zn2+ treatment). (c) DNAzyme molecular machines-controlled migration between a T-cell spheroid (chain-1/chain-3 modified) and a PANC-1 cancer cell spheroid (chain-2 modified). (d) Fluorescent images showing the interaction between a T-cell spheroid and a PANC-1 cancer cell spheroid (approaching each other, forming a linkage, and disconnecting under Zn2+ treatment). BF: bight-field, TAMRA: red fluorescence, AF 488: green fluorescence, Merge: mixed green and red channel. (e) Directional migration of a T-cell spheroid on a cancer cell layer controlled by DNAzyme molecular machines. (f) Bright-field images showing a T-cell spheroid migrating by rolling on a cancer cell layer.
Next, the assembly and disassembly between two cell spheroids (a cancer cell spheroid modified with chain-2 and a T-cell spheroid modified with chain-1/chain-3) were tested (Figure 2c). As shown in Figure 2d, the T-cell and cancer cell spheroids were observed to be linked after encountering. After adding Zn2+ ions, the connected double spheroid assembly was disassociated, showing the feasibility of DNAzyme molecular machines when applied to cell spheroids (Video S5). As a control, T-cell spheroid and cancer cell spheroid without any modification were also assessed, and no connections could be observed within 30 min (Figure S29, Video S6).
Finally, the external interaction control between a T-cell spheroid (modified with chain-1 and chain-3) and a single layer of cancer cells (modified with chain-2) was tested (Figure 2e). As shown in Figure 2f, the T-cell spheroid could be linked to the cancer cell layer after approaching. After Zn2+ ions were added, the T-cell spheroid was disconnected from the bottom and then linked to another location on the cancer cell monolayer (Video S7). Control experiments (a T-cell spheroid above a cancer cell layer without any modification) showed that no linkages could be observed within 30 min (Video S8). Another control experiment by adding PBS showed no signs of disconnection (a T-cell spheroid modified with chain-1 and chain-3 linked with a single layer of cancer cells modified with chain-2), which ruled out the possibility that the disconnection of T-cell spheroids was caused by the liquid flow (Video S9). The above results confirmed that the DNAzyme molecular machines enabled point-to-point migration of T-cell spheroids by rolling on the cancer cell layer, indicating potentials of our design for cancer therapy, as T-cells can be controlled to link cancer cells at a certain location and then leave for targets at other places.
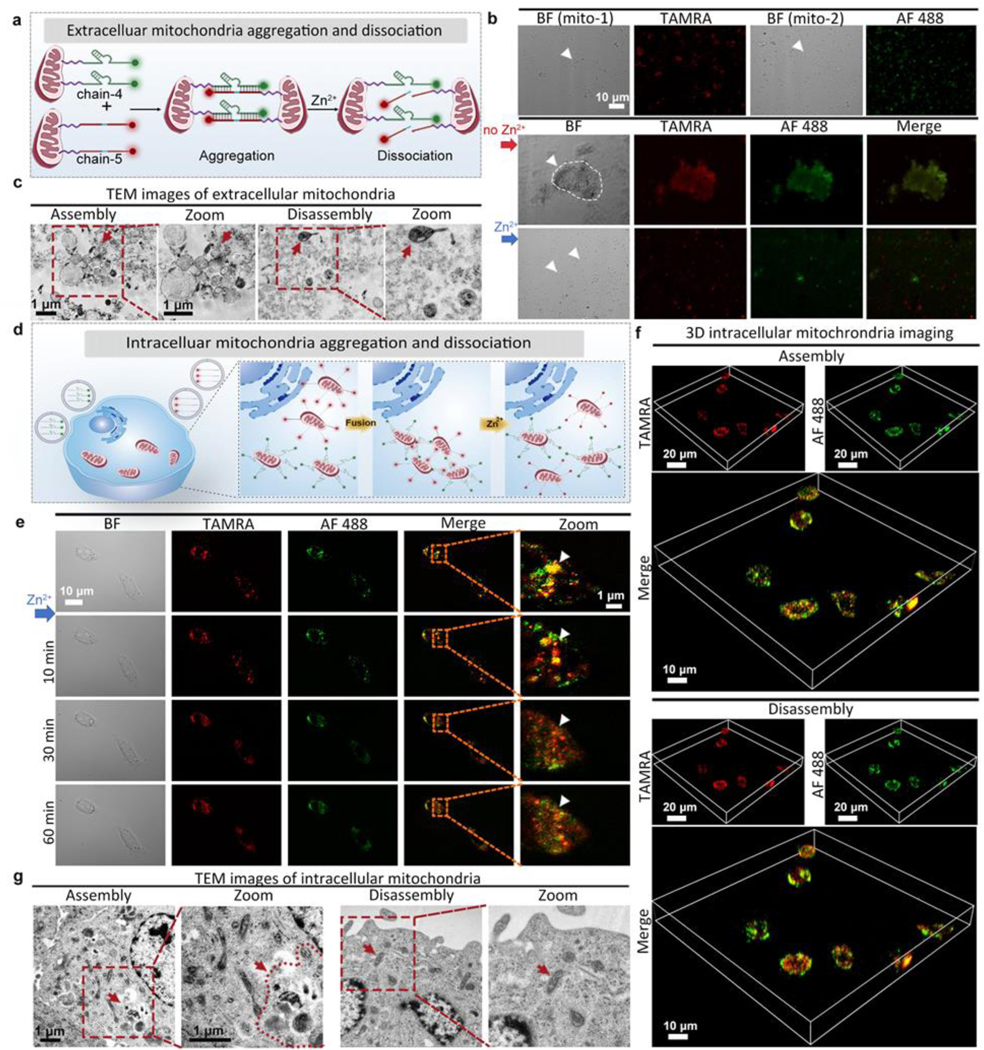
For internal regulation of mitochondria in cancer cells, where the dispersion of mitochondria leads to cancer invasion, a DNAzyme molecular machine (composed by chain-4 and chain-5, Table S1) engineered with mitochondria location peptides is delivered into cancer cells to induce mitochondria aggregation, leading to overproduction of ROS and cause cancer cell death. This aggregation can be disassembled via specific metal ions. Here the Zn2+ induced substrate cleavage was confirmed by gel electrophoresis (Figure S30). Before cell experiments, the interaction of mitochondria was investigated extracellularly. As shown in Figure 3a, the mitochondria extracted from living cells were divided into two groups, and their surface was linked with chain-4 (a Zn2+-specific DNAzyme tagged with AF 488) and chain-5 (a substrate strand tagged with TAMRA), respectively. When these two groups of mitochondria were mixed, mitochondrial aggregation could be formed. As shown in Figure 3b, mitochondrial assemblies could be observed clearly under confocal microscope images. After adding Zn2+ ions, substrate chain-5 was cleaved, causing immediate disconnection within 30 min. TEM images also confirmed the DNAzyme controlled assembly and disassembly of mitochondria. (Figure 3c). Afterwards, the intracellular mitochondrial interactions were tested in living cells (Figure 3d). To improve transfection efficiency, chain-4 and chain-5 were wrapped into liposomes respectively, and then incubated with PANC-1 cells for intracellular delivery (Figure S31). As shown in Figure 3e, when PANC-1 cells were treated with chain-4 (2 μL, 100 μM) for 1 h, and subsequently treated with chain-5 (2 μL, 100 μM) for an additional 1 h, extensive mitochondrial aggregation were observed. After different incubation times (0 min-90 min), the cells were sent for flow cytometry analysis, and the fluorescence intensity reached the maximum after 60 min incubation (Figure S32), which was in accordance with the confocal images (Figure 3e). For quantitative evaluation of chain-4 and chain-5 in PANC-1 cells, the loading efficiency of chain-4 and chain-5 in liposomes was estimated to be 50.0% and 52.6%. (Figure S33, Table S3). After adding Zn2+ ions, mitochondrial disconnection was clearly observed after 30 min under both red and green fluorescent channels. 3D cell images (Figure 3f) and TEM images (Figure 3g) further confirmed the aggregation of intracellular mitochondria under the treatment of chain-4 and chain-5. To confirm that such mitochondrial aggregation was driven by DNAzyme hybridization instead of other nonspecific interactions, control experiments testing extracellular mitochondria or mitochondria in PANC-1 cells without any modification did not show aggregation under TEM images (Figure S34–35). To test the mitochondrial targeting ability, Mito-Tracker Red were used to stain PANC-1 cells treated with chain-4. The fluorescence of Mito-Tracker and chain-4 was highly overlapped, indicating good mitochondrial targeting effect of chains modified mitochondria location peptides (Figure S36). To further test whether the mitochondrial aggregation was realized by the hybridization of chain-4 and chain-5, PANC-1 cells were treated with only chain-4 and incubated for 1 h. As shown in Figure S37, no assembly was observed, with neglectable fluorescence change, which confirmed the hybridization induced aggregation. When these two chains were incubated with HeLa cells and HEK293 cells (with intracellular free zinc concentration higher than PANC-1 cells, as verified by zinc indicator), the aggregation degree of mitochondria was not so obvious as that in zinc deficient PANC-1 cells (Figure S38–39).[29] The above results suggested that the degree of mitochondrial aggregation is associated with the intracellular zinc concentration, as mitochondrial aggregation is more likely to occur in zinc deficient cancer cells instead of normal cells, which is favorable for precise cancer treatment.
Figure 3.
Dynamic intracellular regulation. (a) Zn2+-specific DNAzyme controlled extracellular mitochondrial aggregation and dissociation (group-1: chain-4 modified, group-2: chain-5 modified). (b) Cell images showing the controlled interaction of group-1 and group-2 extracellular mitochondria (1:1). BF: bight-field, AF 488: green fluorescence, TAMRA: red fluorescence, Merge: mixed green and red channel. (c) TEM images of extracellular mitochondria. Left: assembled mitochondria. Right: disassembled mitochondria after adding Zn2+ ions. (d) Internal regulation of intracellular mitochondria interactions by DNAzyme molecular machines (chain-4 and chain-5, delivered by liposomes). (e) Fluorescent images (with squared areas zoomed) showing PANC-1 cells treated with liposomes containing chain-4 and chain-5 for 1 h, followed by observation under different times. BF: bight-field, AF 488: green fluorescence, TAMRA: red fluorescence, Merge: mixed green and red channel. (f) 3D cell images showing assembled mitochondria. (g) TEM images of intracellular mitochondria. Left: assembled mitochondria in PANC-1 cells treated with liposomes containing chain-4 and chain-5. Right: disassembled mitochondria in PANC-1 cells after adding Zn2+ ions.
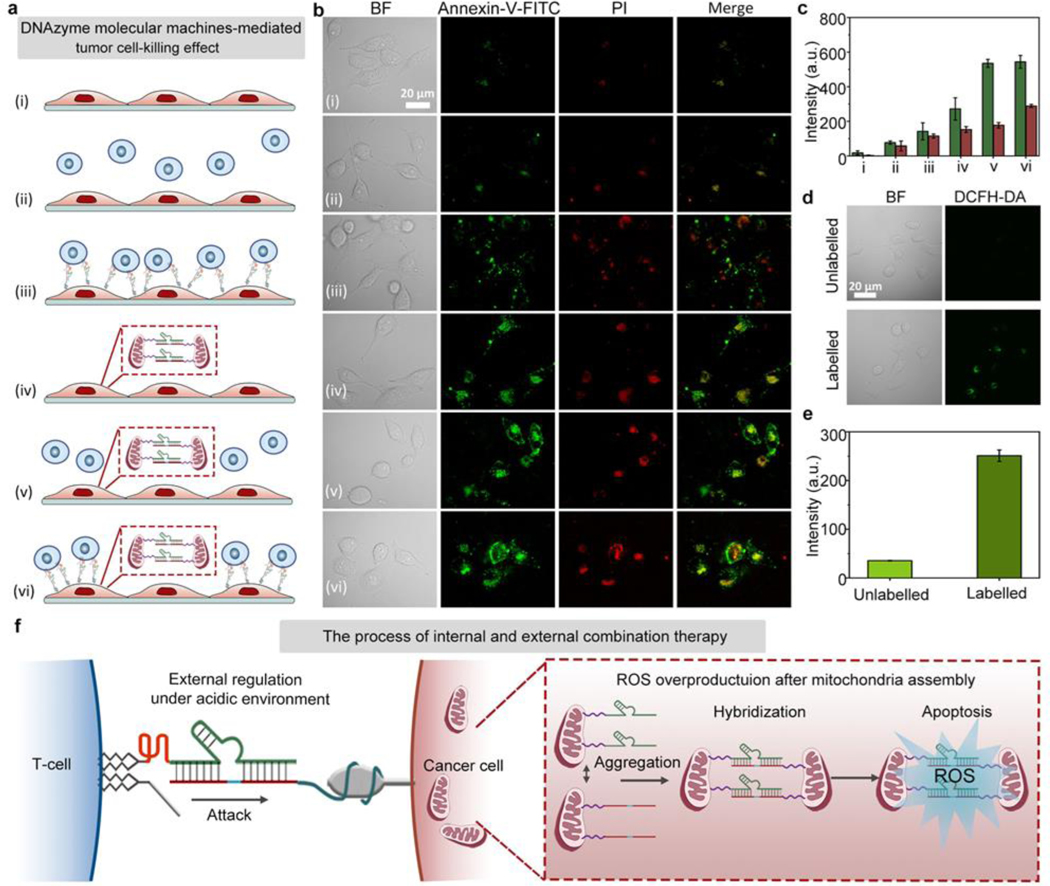
Next, the established combination therapy has been tested in zinc deficient PANC-1 cells under acidic environment. As shown in Figure 4a, the surviving cancer cells were examined by Annexin-V-FITC/PI staining for determining the cancer cell-killing effect by the following different treatment conditions: (i) control PANC-1 cells without any treatment, (ii) PANC-1 cells mixed with unmodified T-cells, (iii) chain-7 (chain-2 without fluorescence marker) modified PANC-1 cells mixed with chain-6 (chain-1 without fluorescence marker)/chain-8 (chain-3 without quencher) modified T-cells (DNAzyme molecular machines for external control of T-cell/cancer cell interactions), (iv) PANC-1 cells mixed with liposome wrapped chain-9 (chain-4 without fluorescence marker)/chain-10 (chain-5 without fluorescence marker, DNAzyme molecular machines for internal control of mitochondrial aggregation), (v) PANC-1 cells mixed with liposome wrapped chain-9/chain-10, followed by incubation with unmodified T-cells, (vi) chain-7 modified PANC-1 cells mixed with liposome wrapped chain-9/chain-10, followed by incubation with chain-6/chain-8 modified T-cells (combination therapy). Afterwards, the apoptosis extent of cancer cells was examined by Annexin-V-FITC/PI staining (Figure 4b). From the microscopy images, it could be seen that the apoptosis fluorescent intensity was highest in cells under combination treatment with both the external and internal regulation by DNAzyme molecular machines. Corresponding fluorescence intensity in Figure 4b further confirmed the above results (Figure 4c). It is worthy to note that PANC-1 cells treated with internal mitochondria modification showed the overproduction of intracellular ROS, contributing to the enhanced cancer cell-killing effect (Figure 4d–e). As shown in Figure S40, the ROS fluorescent intensity in HEK293 cells was neglectable both before and after treatment, indicating no significant effect on cellular viability or mitochondria state of HEK293 cells upon internal mitochondria modification. To further explore the location of intracellular ROS produced upon mitochondria aggregation, commercial tracker dyes including Mito Tracker Red (for targeting mitochondria) and DCFH-DA (for marking ROS) were used to stain PANC-1 cells with internal control of mitochondrial aggregation (Figure S41), and the results showed that the produced ROS was located in mitochondria.[30] Models illustrating the networks were given in Figure 4f, revealing the tendency for mutual regulation between internal and external combination therapy. Compared to control experiments under neutral pH (Figure S42–43), the treatment effect of the combination therapy under acidic environment was much better, with a higher degree of cancer cell-killing. The above results demonstrated the effective design of our strategy by shortening the intercellular distance under acidic pH. As shown in Figure S44, a lower survival rate was observed in PANC-1 cells, while HEK293 cells remained unaffected. Furthermore, the apoptosis fluorescent intensity in HEK293 cells was obviously lower than that in zinc deficient PANC-1 cells under acidic environment (Figure S45–46), indicating the effective design of our strategy. The cell viability experiments showed that the PANC-1 cells remained active after 1 mM Zn2+ treatment of 2 h (Figure S47), excluding the toxicity of zinc ions. These results demonstrated an enhanced synergistic tumor cell-killing effect of the established combination therapy.
Figure 4.
Combination therapy with DNAzyme molecular machines. (a) Combination cancer cell-killing by DNAzyme molecular machines. From (i) to (vi): (i) control, (ii) PANC-1 cells mixed with unmodified T-cells, (iii) chain-7 modified PANC-1 cells mixed with chain-6/chain-8 modified T-cells (DNAzyme molecular machines for external control of T-cell / cancer cell interactions), (iv) PANC-1 cells mixed with liposome wrapped chain-9/chain-10 (DNAzyme molecular machines for internal control of mitochondrial aggregation), (v) PANC-1 cells mixed with unmodified T-cells and liposome wrapped chain-9/chain-10, (vi) chain-7 modified PANC-1 cells mixed with chain-6/chain-8 modified T-cells and liposome wrapped chain-9/chain-10 (combination therapy). (b) Apoptosis fluorescent images showing PANC-1 cells treated with different methods, corresponding to (i) to (vi) in (a). (c) FL intensity comparison histogram of cancer cell-killing under different treatment in (b). (d) Confocal images showing relative ROS levels in unlabeled cancer cells versus labeled cancer cells. (e) Quantified fluorescence intensities corresponding to cell fluorescent images in (d). (f) Illustration showing the combination therapy procedure. BF: bight-field, Annexin-V-FITC: green fluorescence, PI: red fluorescence, Merge: mixed green and red channel.
Conclusion
In this study, we have developed a combination cancer therapy based on molecular machines incorporating engineered DNAzymes and DNA aptamers to induce cancer cell apoptosis from both intercellular regulation of T-cell/cancer cell interactions and intracellular regulation of mitochondrial aggregation. Compared to the conventional recognition between T-cells and cancer cells by using large natural or artificial structures, one major advantage of our strategy is the dynamic regulation of T-cell/cancer cell interactions. To achieve the intercellular external regulation, we designed a DNAzyme molecular machine engineered with aptamer and i-motif to sense and response to the acidic tumor microenvironment. Using the tumor marker MUC-1 as the target protein, the aptamer allows the specific recognition of MUC-1 on cancer cells. The acidic microenvironment of the cancer cells triggers the folding of i-motif sequence, shortening the intercellular distance to induce the killing of cancer cells. As a result, T-cells can be released by metal ion activated DNAzyme-based cleavage. Another advantage of our strategy is the internal control of mitochondrial aggregation in cancer cells. To further induce cancer cell apoptosis, a DNAzyme molecular machine engineered with mitochondria location peptides is delivered into cancer cells to induce mitochondria aggregation, generating excess amount of toxic ROS. Using PANC-1 cells as an example, we demonstrate, for the first time, that such a combination therapy shows an improved therapeutic effect for cancer treatment. Our study demonstrates a powerful strategy based on DNAzymes for the tumor microenvironment-responsive control of extracellular and intracellular cell behaviors including cell recognition, cell-cell approaching, post-treatment separation, and mitochondria aggregation. It not only offers an effective approach for regulating cell-cell interactions, but also provides a powerful cancer therapy that is robust, targeted, and nontoxic to normal cells.
Supplementary Material
Acknowledgements
This research was supported by the National Natural Science Foundation of China (21977031), the U.S. National Institutes of Health (GM141931), Science and Technology Commission of Shanghai Municipality (2018SHZDZX03), Shanghai Science and Technology Committee (22ZR1416800, 19ZR1472300), and the Fundamental Research Funds for the Central Universities. The authors thank Dr. Bo-Hao Yu at Research Center of Analysis and Test of East China University of Science and Technology for the help on laser scanning confocal microscopy analysis.
References
- [1].a) Kwon S, Ko H, You DG, Kataoka K, Park JH, Accounts Chem. Res 2019, 52, 1771–1782. [DOI] [PubMed] [Google Scholar]; b) Wu X, Wu J, Dai J, Chen B, Chen Z, Wang S, Wu F, Lou X, Xia F, Natl. Sci. Rev 2021, 8, nwaa306. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang X, Liu R, Zhu W, Chu H, Yu H, Wei P, Wu X, Zhu H, Gao H, Liang J, Li G, Yang W, Nature 2019, 571, 127–131. [DOI] [PubMed] [Google Scholar]; d) Dong J, Ouyang Y, Wang J, O’Hagan MP, Willner I, ACS Nano 2022. DOI: 10.1021/acsnano.1c11631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Argyriou AA, Bruna J, Marmiroli P, Cavaletti G, Crit. Rev. Oncol./Hematol 2012, 82, 51–77. [DOI] [PubMed] [Google Scholar]; b) Monsuez JJ, Charniot JC, Vignat N, Artigou JY, Int. J. Cardiol 2010, 144, 3–15. [DOI] [PubMed] [Google Scholar]
- [3].Powerful Combination Therapies. Nat. Biomed. Eng 2018, 2, 555–556. [DOI] [PubMed] [Google Scholar]
- [4].a) Hu CJ, Zhang L, Biochem. Pharmacol. 2012, 83, 1104–1111. [DOI] [PubMed] [Google Scholar]; b) Qi J, Jin F, Xu X, Du Y, Int. J. Nanomed 2021, 16, 1435–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Pusuluri A, Wu D, Mitragotri S, J. Control Release 2019, 305, 130–154. [DOI] [PubMed] [Google Scholar]
- [5].a) Patel SA, Minn AJ, Immunity 2018, 48, 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A, Cell 2017, 168, 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF, Sci. Transl. Med 2013, 5, 200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Zhao Z, Ukidve A, Kim j., Mitragotri S, Cell 2020, 181, 151–167. [DOI] [PubMed] [Google Scholar]; b) Zhou Y, Chen X, Cao J, Gao H, J. Mat. Chem. B 2020, 8, 6765–6781. [DOI] [PubMed] [Google Scholar]; c) Blanco E, Shen H, Ferrari M, Nat. Biotechnol 2015, 33, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Ritter A, Asano Y, Stinchcombe J, Dieckmann NMG, Chen BC, Gawden-Bone C, Engelenburg S, Legant W, Gao L, Davidson MW, Betzig E, Lippincott-Schwartz J, Griffiths GM, Immunity 2015, 42, 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cardle II, Cheng EL, Jensen MC, Pun SH, Accounts Chem. Res 2020, 53, 1724–1738. [DOI] [PubMed] [Google Scholar]
- [8].a) Zorov DB, Filburn CR, Klotz LO, Zweier JL, Sollott SJ, J. Exp. Med 2000, 192, 1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zorov DB, Juhaszova M, Sollott SJ, Physiol. Rev 2014, 94, 909–950. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Park J, Lee J, Choi C, PLoS One 2011, 6, e23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Waldman AD, Fritz JM, Lenardo MJ, Nat. Rev. Immunol 2020, 20, 651–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) A Desai SP, Bhatia SN, Toner M, Irimia D, Biophys. J 2013, 104, 2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Caino MC, Ghosh JC, Chae YC, Vaira V, Rivadeneira DB, Faversani A, Rampini P, Kossenkov AV, Aird KM, Zhang R, Proc. Natl. Acad. Sci. U. S. A 2015, 112, 8638–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Onodera Y, Nam JM, Horikawa M, Shirato H, Sabe H, Nat. Commun 2018, 9, 2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Hwang K, Hosseinzadeh P, Lu Y, Inorg. Chim. Acta 2016, 452, 12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li Y, Breaker RR, Curr. Opin. Struct. Biol 1999, 9, 315–323. [DOI] [PubMed] [Google Scholar]; c) Hwang K, Mou Q, Lake RJ, Xiong M, Holland B, Lu Y, Inorg. Chem 2019, 58, 13696−13708. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Liu J, Lu Y, J. Am. Chem. Soc 2002, 124, 15208–15216. [DOI] [PubMed] [Google Scholar]
- [13].a) Alam S, Kelleher SL, Nutrients. 2012, 4, 875–903. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pan Z, Choi S, Ouadid-Ahidouch H, Yang JM, Beattie JH, Korichneva I, Front Biosci. 2017, 22, 623–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Qian R, Zhou Z, Guo W, Wu Y, Yang Z, Lu Y, J. Am. Chem. Soc 2021, 143, 5737–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li J, Zheng W, Kwon AH, Lu Y, Nucleic Acids Res. 2000, 28, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].a) Chung M, Koo BJ, Boxer SG, Faraday Discuss. 2013, 161, 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) You M, Lyu Y, Han D, Qiu L, Liu Q, Chen T, Wu CS, Peng L, Zhang L, Bao G, Tan W, Nat. Nanotechnol 2017, 12, 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chan Y, Lengerich BV, Boxer SG, Biointerphases 2008, 3, FA17-FA21. [DOI] [PubMed] [Google Scholar]; d) J Woods EC, Yee NA, Shen J, Bertozzi CR, Angew. Chem.-Int. Edit 2015, 54, 15782–15788. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Delaverisa CS, Webstera ER, Banika SM, Boxera SG, Bertozz CR, Proc. Natl. Acad. Sci. U. S. A 2020, 17, 12643−12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Zeng S, Liu D, Li C, Yu F, Fan L, Lei C, Huang Y, Nie Z, Yao S, Anal. Chem 2018, 90, 13459–13466. [DOI] [PubMed] [Google Scholar]; b) Ying L, Xie N, Yang Y, Yang X, Zhou Q, Yin B, Huang J, Wang K, Chem. Commun 2016, 52, 7818–7821. [DOI] [PubMed] [Google Scholar]
- [18].a) Zhang J, Qiu Z, Fan J, He F, Kang W, Yang S, Wang HH, Huang J, Nie Z, Angew. Chem.-Int. Edit 2021, 60, 6733–6743. [DOI] [PubMed] [Google Scholar]; b) Lv J, Chang S, Wang XY, Zhou ZR, Chen BB, Qian RC, Li DW, Sens. Actuator B-Chem 2022, 351, 130877. [Google Scholar]
- [19].a) Sun S, Ning X, Zhang G, Wang YC, Peng C, Zheng J. Angew. Chem.-Int. Edit 2016, 55, 2421–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang LX, Tan HC, Hanson BJ, Ooi EE. J. Virol. Methods 2010, 167, 172–177. [DOI] [PubMed] [Google Scholar]
- [20].Keshri P, Zhao B, Xie T, Bagheri Y, Chambers J, Sun Y, You M, Angew. Chem.-Int. Edit 2021, 60, 15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Boedtkjer E, Pedersen SF, Annu. Rev. Physiol 2020, 82, 21.1–21.24. [DOI] [PubMed] [Google Scholar]
- [22].Gold DV, Modrak DE, Newsome G, Karacay H, Sharkey RM, Goldenberg DM, J. Clin. Oncol 2006, 24, 4096–4096. [Google Scholar]
- [23].a) Ferreira CSM, Matthews CS, and Missailidis S. Tumor. Biol 2006, 27, 289–301. [DOI] [PubMed] [Google Scholar]; b) Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 15416–15421. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nabavinia MS, Gholoobi A, Charbgoo F, Nabavinia M, Ramezani M, Abnous K. Med Res Rev. 2017, 37, 1518–1539. [DOI] [PubMed] [Google Scholar]
- [24].Li X, Duan X, Yang P, Li L, Tzang B, Anal. Chem 2021, 93, 4059–4065. [DOI] [PubMed] [Google Scholar]
- [25].Costello LC, Zou J, Desouki MM, Franklin RB, Gastrointest J. Cancer 2012, 43, 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].a) Kambe T, Hashimoto A, Fujimoto S, Cell Mol Life Sci. 2014, 71, 3281–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) John E, Laskow TC, Buchser WJ, Pitt BR, Basse PH, Butterfield LH, Kalinski P, J Transl Med. 2010, 8, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Boedtkjer E, Pedersen SF, Annu. Rev. Physiol 2020, 82, 103–126. [DOI] [PubMed] [Google Scholar]
- [28].Yang Z, Loh KY, Chu YT, Feng R, Satyavolu NSR, Xiong M, Nakamata Huynh SM, Hwang K, Li L, Xing H, Zhang X, Chemla YR, Gruebele M, Lu Y, J. Am. Chem. Soc 2018, 140, 17656−17665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].a) Franklin RB, Zou J, Costello LC, Cancer Biol. Ther 2014, 15, 1431–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Costello LC, Zou J, Desouki MM, Franklin RB, Gastrointest J. Cancer 2012, 43, 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].a) Bandy B, Davison AJ, Free Radic. Biol. Med 1990, 8, 523–539. [DOI] [PubMed] [Google Scholar]; b) Indo HP, Davidson M, Yen HC, Suenaga S, Tomita K, Nishii T, Higuchi M, Koga Y, Ozawa T, Majima HJ, Mitochondrion 2007, 7, 106–118. [DOI] [PubMed] [Google Scholar]; c) Majima H, Oberley TD, Furukawa K, Mattson MP, Yen HC, Szweda LI, St DK. Clair, J. Biol. Chem 1998, 273, 8217–8224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.