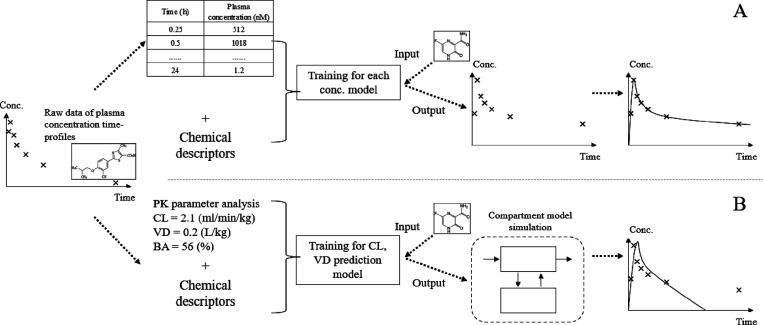
Figure 1.
Overview of the workflow of this research. (A) We developed models to predict in-vivo concentration data using the compound structure (MACCS Keys) and both in silico and in vitro PK parameters. Separate models were built for each time point after compound administration. The predictions were subsequently combined to form a predicted time–concentration curve (see the section “Explanatory Variables” in “Materials and Method”). (B) The conventional compartmental model method to predict compound plasma concentration–time profiles (see the section “Baseline Compartmental Model” in “Materials and Method”). In the conventional method, CL and VD needed to be predicted, which was achieved using machine learning models built in this study, and then used as inputs of the compartmental model. Finally, plasma concentration–time profiles were obtained.