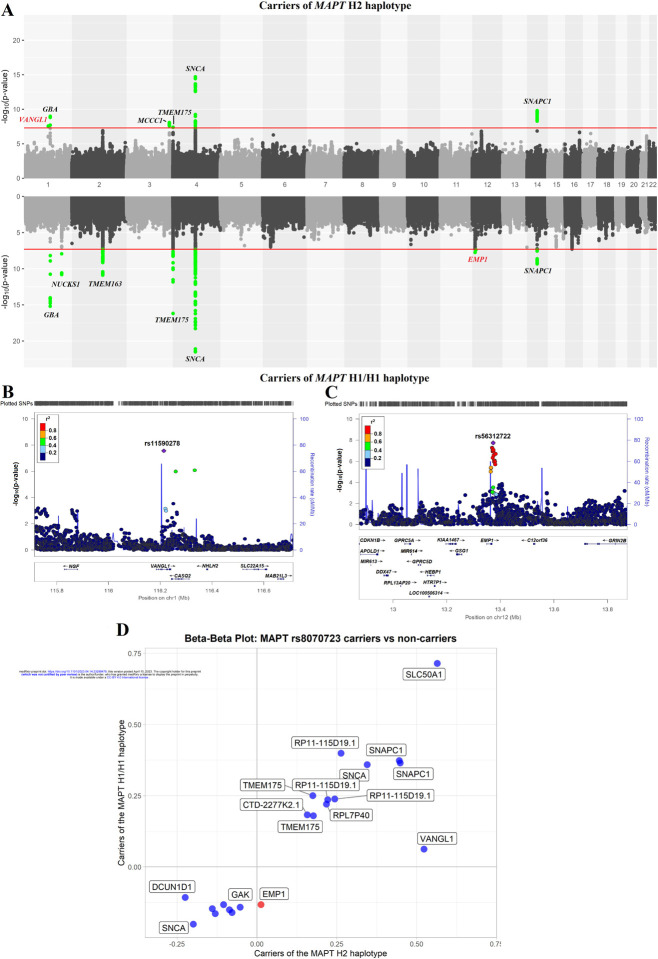
Figure 1.
A. Miami plot of carriers (top) and non-carriers (bottom) of MAPT H2 haplotype of selected variants. The red line indicates GWAS significance threshold. Green dots indicate passing the threshold variants. New loci with potential interaction with MAPT highlighted in red, other loci highlighted in black. B. Locus zoom plot of VANGL1 variants (+/− 500kb) in the stratified PD risk GWAS of the MAPT H2 haplotype carriers. C. Locus zoom plot of EMP1 variants (+/− 500kb) in the stratified PD risk GWAS of the MAPT H1/H1 haplotype carriers. The position of the variants on the chromosome (x axis) is plotted against the log10-scaled p-values (left y axis). SNP with the smallest p-value is indicated by a purple diamond. Linkage disequilibrium scores related to the top variant are defined by different colors, explained by the legend on the upper left corner. The right vertical axis indicates the regional recombination rate (cM/Mb). Abbreviations: chr, chromosome; cM, centimorgan; Mb, Megabase; PD, Parkinson’s disease; SNP, single nucleotide polymorphism. D. Beta-beta plots for independent SNPs that reached genome wide association level of significance in carriers or non-carriers of MAPT rs8070723 (Carriers of the MAPT H2 haplotype vs carriers of the MAPT H1/H1 haplotype). Red circles denote SNPs with effect size (beta) difference of more than 5 times between carriers of MAPT H2 haplotype and carriers of the MAPT H1/H1 haplotype.