Extended Data Fig. 8 |. Meta-analysis of all cohorts using random effects model identifies organisms differentially enriched in melanoma patients treated with anti-PD-1 therapy in separate cohorts by response status.
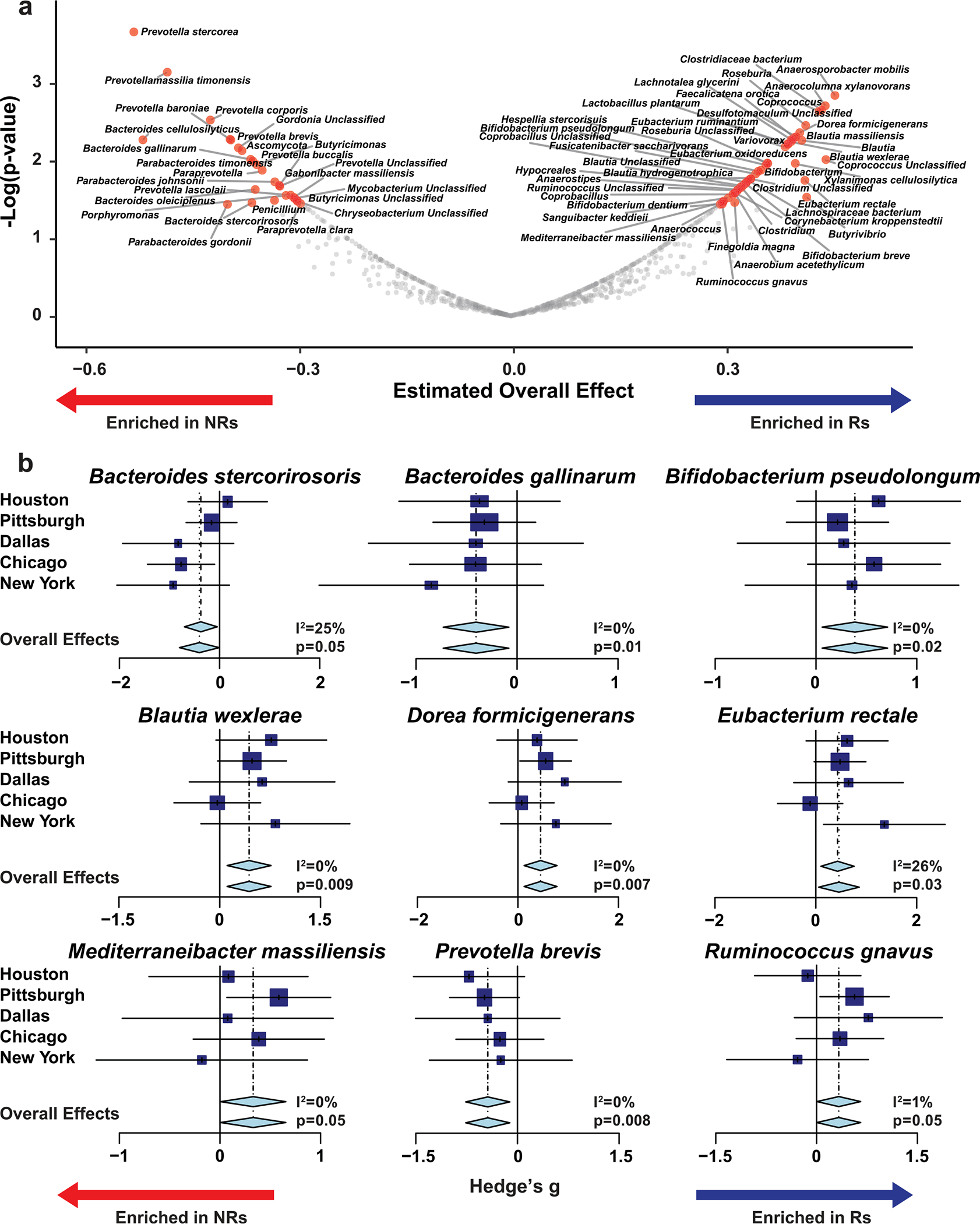
a. Random effect model meta-analysis of differentially abundant bacteria between responders (Rs) and non-responders (NRs) from five cohorts (n = 185) including Pittsburgh early sample cohort (n = 63) using shotgun metagenomic sequencing. All significant bacterial taxa enriched in Rs and NRs are shown. b. Forest plots depict association of representative bacterial species with response to anti-PD-1 therapy. Within each plot, names of various cohorts are shown on separate lines, while Hedge’s g (squares, standardized mean differences, size proportional to sample size) and associated 95% confidence intervals (bars) are shown, along with the dotted vertical line of no effect. To control for unobserved heterogeneity, we separately evaluated Hedge’s g using random effect model on metagenomic data and performed I2 test for heterogeneity as shown. P-values were calculated using random effect model.
