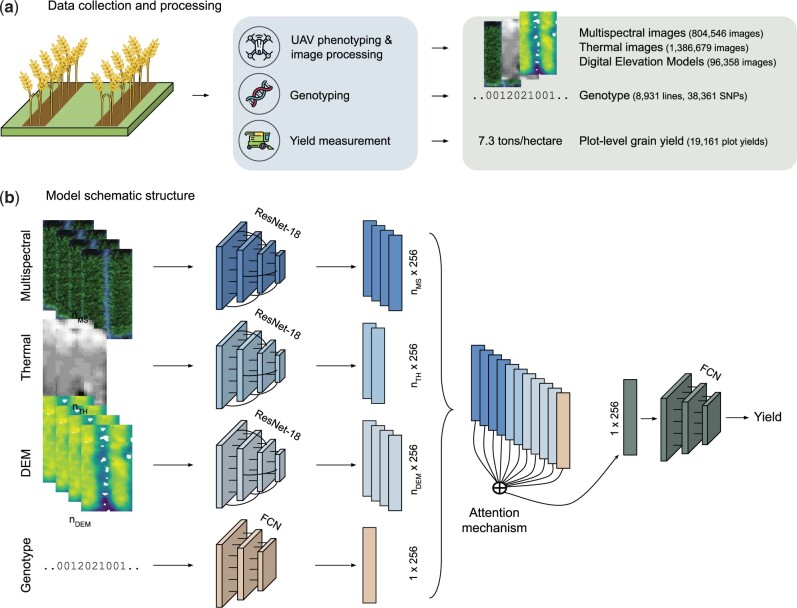
Figure 1.
PheGeMIL leverages multi-channel inputs, combining dynamic phenotypic observations with genotypes. (a) Advanced spring wheat (Triticum aestivum L.) breeding lines from the International Maize and Wheat Improvement Center (CIMMYT) breeding program were sown during season 2017–2018. Multispectral and thermal images were collected by UAV flights over the experiment field and digital elevation model (DEM) developed from the images. Lines were profiled using genotyping-by-sequencing with calling of single nucleotide variants (SNPs). Final crop yield was measured by combine harvest in tons per hectare (t/ha) on a per plot basis. (b) The model flexibly enables the fusion of information across channels via a single deep learning architecture that is fully differentiable. Encoder networks for each data type transform images or genotypes into vectors of fixed dimensionality (256 dimensions). The attention mechanism then combines these vectorial representations in a single 256-dimensional vector, independently of the number of input images provided. The unique representation is then fed in a final fully connected network to predict plot-level yield. nMS, number of multispectral images for a given sample; nTH, number of thermal images for a given sample; nDEM, number of digital elevation model images for a given sample; ResNet-18, a convolutional residual neural network architecture for image feature extraction; FCN, fully connected neural network architecture; SNPs, single nucleotide polymorphisms.